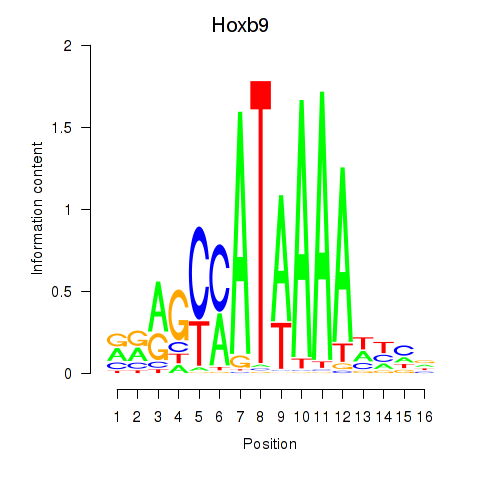
|
chr15_+_6552270
Show fit
|
0.26 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein
|
|
chr7_-_12468931
Show fit
|
0.25 |
ENSMUST00000233304.2
ENSMUST00000233373.2
ENSMUST00000233874.2
ENSMUST00000233902.2
|
Vmn2r56
|
vomeronasal 2, receptor 56
|
|
chr10_+_129153986
Show fit
|
0.20 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780
|
|
chr9_+_38119661
Show fit
|
0.19 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893
|
|
chr15_+_3300249
Show fit
|
0.14 |
ENSMUST00000082424.12
ENSMUST00000159158.9
ENSMUST00000159216.10
ENSMUST00000160311.3
|
Selenop
|
selenoprotein P
|
|
chr14_-_33700719
Show fit
|
0.12 |
ENSMUST00000166737.2
|
Zfp488
|
zinc finger protein 488
|
|
chr10_-_40018243
Show fit
|
0.12 |
ENSMUST00000092566.8
ENSMUST00000213488.2
|
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10
|
|
chr19_-_7319157
Show fit
|
0.11 |
ENSMUST00000164205.8
ENSMUST00000165286.8
ENSMUST00000168324.2
ENSMUST00000032557.15
|
Mark2
|
MAP/microtubule affinity regulating kinase 2
|
|
chrX_-_49108236
Show fit
|
0.11 |
ENSMUST00000213556.3
ENSMUST00000213463.2
|
Olfr1323
|
olfactory receptor 1323
|
|
chr7_-_12342715
Show fit
|
0.11 |
ENSMUST00000233892.2
|
Vmn2r53
|
vomeronasal 2, receptor 53
|
|
chr8_-_5155347
Show fit
|
0.11 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2
|
|
chr2_-_36752671
Show fit
|
0.11 |
ENSMUST00000213676.2
ENSMUST00000215137.2
|
Olfr351
|
olfactory receptor 351
|
|
chr2_+_85545763
Show fit
|
0.10 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009
|
|
chr2_+_88479038
Show fit
|
0.09 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1
|
|
chr2_-_51039112
Show fit
|
0.09 |
ENSMUST00000154545.2
ENSMUST00000017288.9
|
Rnd3
|
Rho family GTPase 3
|
|
chr7_+_110367375
Show fit
|
0.08 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr13_-_22689551
Show fit
|
0.08 |
ENSMUST00000228020.2
|
Vmn1r202
|
vomeronasal 1 receptor 202
|
|
chr11_-_109188917
Show fit
|
0.08 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9
|
|
chr1_+_59296065
Show fit
|
0.07 |
ENSMUST00000160662.8
ENSMUST00000114248.3
|
Cdk15
|
cyclin-dependent kinase 15
|
|
chr2_+_22512195
Show fit
|
0.07 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2
|
|
chr19_+_13208692
Show fit
|
0.07 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463
|
|
chr17_-_29226886
Show fit
|
0.06 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38
|
|
chr4_+_115594951
Show fit
|
0.06 |
ENSMUST00000106522.9
|
Efcab14
|
EF-hand calcium binding domain 14
|
|
chr10_+_111342147
Show fit
|
0.06 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1
|
|
chr10_+_3822667
Show fit
|
0.06 |
ENSMUST00000136671.8
ENSMUST00000042438.13
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr3_+_66892979
Show fit
|
0.05 |
ENSMUST00000162362.8
ENSMUST00000065074.14
ENSMUST00000065047.13
|
Rsrc1
|
arginine/serine-rich coiled-coil 1
|
|
chr17_+_33857030
Show fit
|
0.05 |
ENSMUST00000052079.8
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1
|
|
chrX_+_158623460
Show fit
|
0.05 |
ENSMUST00000112451.8
ENSMUST00000112453.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr4_-_119217079
Show fit
|
0.04 |
ENSMUST00000143494.3
ENSMUST00000154606.9
|
Ccdc30
|
coiled-coil domain containing 30
|
|
chr5_+_137786031
Show fit
|
0.04 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1
|
|
chr18_+_38809771
Show fit
|
0.04 |
ENSMUST00000134388.2
ENSMUST00000148850.8
|
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene
Rho GTPase activating protein 26
|
|
chr9_+_38259707
Show fit
|
0.04 |
ENSMUST00000217063.2
|
Olfr898
|
olfactory receptor 898
|
|
chr17_+_38456172
Show fit
|
0.04 |
ENSMUST00000215078.3
ENSMUST00000215549.3
ENSMUST00000173610.2
|
Olfr133
|
olfactory receptor 133
|
|
chr4_+_115594918
Show fit
|
0.04 |
ENSMUST00000106525.9
|
Efcab14
|
EF-hand calcium binding domain 14
|
|
chr5_+_63969706
Show fit
|
0.04 |
ENSMUST00000081747.8
ENSMUST00000196575.5
|
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene
|
|
chr17_+_18801989
Show fit
|
0.04 |
ENSMUST00000165692.8
|
Vmn2r96
|
vomeronasal 2, receptor 96
|
|
chr8_-_70591746
Show fit
|
0.04 |
ENSMUST00000212320.2
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein
|
|
chr2_-_32976378
Show fit
|
0.04 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3
|
|
chr19_-_7319211
Show fit
|
0.04 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2
|
|
chr2_-_88768449
Show fit
|
0.04 |
ENSMUST00000215205.2
ENSMUST00000213412.2
|
Olfr1211
|
olfactory receptor 1211
|
|
chr14_+_20724378
Show fit
|
0.04 |
ENSMUST00000224492.2
ENSMUST00000223751.2
ENSMUST00000225108.2
ENSMUST00000224754.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae)
|
|
chr2_+_111984716
Show fit
|
0.04 |
ENSMUST00000214063.2
ENSMUST00000217533.2
|
Olfr1318
|
olfactory receptor 1318
|
|
chr9_-_40004854
Show fit
|
0.04 |
ENSMUST00000050996.6
ENSMUST00000213087.4
|
Olfr983
|
olfactory receptor 983
|
|
chr14_-_47426863
Show fit
|
0.03 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1
|
|
chr8_-_70591799
Show fit
|
0.03 |
ENSMUST00000075724.9
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein
|
|
chr11_+_50905063
Show fit
|
0.03 |
ENSMUST00000217480.2
ENSMUST00000215409.2
|
Olfr54
|
olfactory receptor 54
|
|
chr9_-_101128976
Show fit
|
0.03 |
ENSMUST00000075941.12
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr2_-_45002902
Show fit
|
0.03 |
ENSMUST00000076836.13
ENSMUST00000176732.8
ENSMUST00000200844.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_79394904
Show fit
|
0.03 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr7_+_43339842
Show fit
|
0.03 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14
|
|
chr7_+_44545501
Show fit
|
0.03 |
ENSMUST00000071207.14
ENSMUST00000209132.2
ENSMUST00000207069.2
ENSMUST00000209039.2
ENSMUST00000207939.3
ENSMUST00000207485.2
ENSMUST00000208179.2
|
Fuz
|
fuzzy planar cell polarity protein
|
|
chr17_-_29226965
Show fit
|
0.03 |
ENSMUST00000009138.13
ENSMUST00000119274.3
|
Stk38
|
serine/threonine kinase 38
|
|
chr2_-_32977182
Show fit
|
0.03 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3
|
|
chr9_-_101076198
Show fit
|
0.03 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr13_+_83723743
Show fit
|
0.03 |
ENSMUST00000198217.5
ENSMUST00000199210.5
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr17_-_29226700
Show fit
|
0.03 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38
|
|
chr7_-_44711771
Show fit
|
0.03 |
ENSMUST00000210101.2
ENSMUST00000209219.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2
|
|
chr8_-_112718888
Show fit
|
0.03 |
ENSMUST00000034427.12
ENSMUST00000139820.8
|
Adat1
|
adenosine deaminase, tRNA-specific 1
|
|
chr2_+_29509704
Show fit
|
0.03 |
ENSMUST00000095087.11
ENSMUST00000091146.12
ENSMUST00000102872.11
ENSMUST00000147755.10
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr16_-_17348996
Show fit
|
0.03 |
ENSMUST00000100125.12
|
Thap7
|
THAP domain containing 7
|
|
chr16_-_17348882
Show fit
|
0.03 |
ENSMUST00000231548.2
ENSMUST00000232041.2
ENSMUST00000231288.2
|
Thap7
|
THAP domain containing 7
|
|
chr7_-_104677667
Show fit
|
0.03 |
ENSMUST00000215899.2
ENSMUST00000214318.3
|
Olfr675
|
olfactory receptor 675
|
|
chr6_+_29694181
Show fit
|
0.03 |
ENSMUST00000046750.14
ENSMUST00000115250.4
|
Tspan33
|
tetraspanin 33
|
|
chr8_+_121215155
Show fit
|
0.03 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein
|
|
chr16_-_19241884
Show fit
|
0.02 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165
|
|
chr11_-_99134885
Show fit
|
0.02 |
ENSMUST00000103132.10
ENSMUST00000038214.7
|
Krt222
|
keratin 222
|
|
chr2_+_89821818
Show fit
|
0.02 |
ENSMUST00000216953.3
|
Olfr1261
|
olfactory receptor 1261
|
|
chr2_+_89821565
Show fit
|
0.02 |
ENSMUST00000111509.3
ENSMUST00000213909.3
|
Olfr1261
|
olfactory receptor 1261
|
|
chr7_-_12829100
Show fit
|
0.02 |
ENSMUST00000209822.3
ENSMUST00000235753.2
|
Vmn1r85
|
vomeronasal 1 receptor 85
|
|
chr7_-_29643172
Show fit
|
0.02 |
ENSMUST00000108212.8
|
Zfp74
|
zinc finger protein 74
|
|
chr7_+_29931309
Show fit
|
0.02 |
ENSMUST00000019882.16
ENSMUST00000149654.8
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I
|
|
chr18_+_20509771
Show fit
|
0.02 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta
|
|
chr2_+_87436556
Show fit
|
0.02 |
ENSMUST00000213103.3
ENSMUST00000216580.2
|
Olfr1130
|
olfactory receptor 1130
|
|
chr19_+_13628455
Show fit
|
0.02 |
ENSMUST00000213900.2
|
Olfr1490
|
olfactory receptor 1490
|
|
chr4_+_132857816
Show fit
|
0.02 |
ENSMUST00000084241.12
ENSMUST00000138831.2
|
Wasf2
|
WASP family, member 2
|
|
chr9_-_63929308
Show fit
|
0.02 |
ENSMUST00000179458.2
ENSMUST00000041029.6
|
Smad6
|
SMAD family member 6
|
|
chr4_+_145311759
Show fit
|
0.02 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268
|
|
chr6_+_134958681
Show fit
|
0.02 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1
|
|
chr6_-_52190299
Show fit
|
0.02 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308
|
|
chr15_+_81820954
Show fit
|
0.02 |
ENSMUST00000038757.8
ENSMUST00000230633.2
|
Csdc2
|
cold shock domain containing C2, RNA binding
|
|
chr9_+_64142483
Show fit
|
0.02 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4
|
|
chr6_+_70675416
Show fit
|
0.02 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2
|
|
chr10_+_129306867
Show fit
|
0.02 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788
|
|
chr14_+_20724366
Show fit
|
0.02 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae)
|
|
chr5_-_32903687
Show fit
|
0.02 |
ENSMUST00000135248.2
|
Pisd
|
phosphatidylserine decarboxylase
|
|
chrY_-_9132561
Show fit
|
0.01 |
ENSMUST00000171947.3
|
Gm21292
|
predicted gene, 21292
|
|
chr2_-_27354006
Show fit
|
0.01 |
ENSMUST00000164296.8
|
Brd3
|
bromodomain containing 3
|
|
chr2_+_111355548
Show fit
|
0.01 |
ENSMUST00000217845.3
|
Olfr1293-ps
|
olfactory receptor 1293, pseudogene
|
|
chr15_-_41733099
Show fit
|
0.01 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein
|
|
chr7_-_104983311
Show fit
|
0.01 |
ENSMUST00000211006.3
ENSMUST00000216230.2
|
Olfr690
|
olfactory receptor 690
|
|
chr9_-_107648144
Show fit
|
0.01 |
ENSMUST00000183248.3
ENSMUST00000182022.8
ENSMUST00000035199.13
ENSMUST00000182659.8
|
Rbm5
|
RNA binding motif protein 5
|
|
chr2_-_57942844
Show fit
|
0.01 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein
|
|
chr12_+_71184614
Show fit
|
0.01 |
ENSMUST00000045907.16
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene
|
|
chr9_+_121196034
Show fit
|
0.01 |
ENSMUST00000210636.2
ENSMUST00000045903.8
|
Trak1
|
trafficking protein, kinesin binding 1
|
|
chr11_-_78277384
Show fit
|
0.01 |
ENSMUST00000108294.2
|
Foxn1
|
forkhead box N1
|
|
chr17_+_36172210
Show fit
|
0.01 |
ENSMUST00000074259.15
ENSMUST00000174873.2
|
Nrm
|
nurim (nuclear envelope membrane protein)
|
|
chr2_+_36674288
Show fit
|
0.01 |
ENSMUST00000213498.2
ENSMUST00000214909.3
ENSMUST00000215199.2
|
Olfr348
|
olfactory receptor 348
|
|
chr2_+_147206910
Show fit
|
0.01 |
ENSMUST00000109968.3
|
Pax1
|
paired box 1
|
|
chr4_-_58787716
Show fit
|
0.01 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267
|
|
chr6_-_122317484
Show fit
|
0.01 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1
|
|
chr2_-_80277969
Show fit
|
0.01 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein
|
|
chr7_-_85974838
Show fit
|
0.01 |
ENSMUST00000214977.2
|
Olfr308
|
olfactory receptor 308
|
|
chr7_-_37970734
Show fit
|
0.01 |
ENSMUST00000032585.8
|
Pop4
|
processing of precursor 4, ribonuclease P/MRP family, (S. cerevisiae)
|
|
chr6_+_70640233
Show fit
|
0.01 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5
|
|
chr2_+_85804239
Show fit
|
0.01 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029
|
|
chr6_-_122317457
Show fit
|
0.01 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1
|
|
chr6_+_70648743
Show fit
|
0.01 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4
|
|
chr17_+_36172235
Show fit
|
0.01 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein)
|
|
chr9_+_35819708
Show fit
|
0.01 |
ENSMUST00000176049.2
ENSMUST00000176153.2
|
Pate13
|
prostate and testis expressed 13
|
|
chr11_+_115294560
Show fit
|
0.01 |
ENSMUST00000153983.8
ENSMUST00000106539.10
ENSMUST00000103036.5
|
Mrpl58
|
mitochondrial ribosomal protein L58
|
|
chr11_+_49346170
Show fit
|
0.01 |
ENSMUST00000214541.2
|
Olfr1387
|
olfactory receptor 1387
|
|
chr3_+_108498595
Show fit
|
0.01 |
ENSMUST00000051145.15
|
Wdr47
|
WD repeat domain 47
|
|
chr8_-_25164767
Show fit
|
0.00 |
ENSMUST00000033957.12
|
Adam18
|
a disintegrin and metallopeptidase domain 18
|
|
chr5_+_128677863
Show fit
|
0.00 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10
|
|
chr2_+_84818538
Show fit
|
0.00 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3
|
|
chr9_+_45014092
Show fit
|
0.00 |
ENSMUST00000217074.2
|
Jaml
|
junction adhesion molecule like
|
|
chr11_-_58446443
Show fit
|
0.00 |
ENSMUST00000216725.2
ENSMUST00000215717.2
ENSMUST00000108824.3
|
Olfr328
|
olfactory receptor 328
|
|
chr7_-_103191924
Show fit
|
0.00 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612
|
|
chr4_+_44012638
Show fit
|
0.00 |
ENSMUST00000107847.10
ENSMUST00000170241.8
ENSMUST00000107849.10
ENSMUST00000107851.10
ENSMUST00000107845.4
|
Clta
|
clathrin, light polypeptide (Lca)
|
|
chr7_+_55699883
Show fit
|
0.00 |
ENSMUST00000205653.2
ENSMUST00000076226.13
|
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2
|
|
chr6_-_87958611
Show fit
|
0.00 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone
|
|
chr7_+_142086749
Show fit
|
0.00 |
ENSMUST00000038675.7
|
Mrpl23
|
mitochondrial ribosomal protein L23
|
|
chr2_+_86655007
Show fit
|
0.00 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094
|
|
chr12_+_105125949
Show fit
|
0.00 |
ENSMUST00000085043.7
ENSMUST00000222342.2
|
Tcl1b1
|
T cell leukemia/lymphoma 1B, 1
|
|
chr9_+_92191415
Show fit
|
0.00 |
ENSMUST00000150594.8
ENSMUST00000098477.8
|
1700057G04Rik
|
RIKEN cDNA 1700057G04 gene
|
|
chr5_-_26310360
Show fit
|
0.00 |
ENSMUST00000063524.3
|
5031410I06Rik
|
RIKEN cDNA 5031410I06 gene
|
|
chr8_-_4155758
Show fit
|
0.00 |
ENSMUST00000138439.2
ENSMUST00000145007.8
|
Cd209f
|
CD209f antigen
|
|
chr1_+_88139678
Show fit
|
0.00 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr3_+_82933383
Show fit
|
0.00 |
ENSMUST00000029630.15
ENSMUST00000166581.4
|
Fga
|
fibrinogen alpha chain
|
|
chr19_-_45800730
Show fit
|
0.00 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2
|
|
chr4_+_147056433
Show fit
|
0.00 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989
|
|
chr8_-_96302083
Show fit
|
0.00 |
ENSMUST00000213096.2
ENSMUST00000052690.13
|
Prss54
|
protease, serine 54
|
|
chr3_+_137923521
Show fit
|
0.00 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr16_+_38405718
Show fit
|
0.00 |
ENSMUST00000165631.2
|
Tmem39a
|
transmembrane protein 39a
|
|
chr7_-_103661957
Show fit
|
0.00 |
ENSMUST00000106862.3
|
Olfr639
|
olfactory receptor 639
|
|
chr19_-_13075176
Show fit
|
0.00 |
ENSMUST00000208913.2
ENSMUST00000215229.2
|
Olfr1457
|
olfactory receptor 1457
|
|
chrX_+_35664933
Show fit
|
0.00 |
ENSMUST00000115253.3
|
Gm6268
|
predicted gene 6268
|
|
chr14_+_56279498
Show fit
|
0.00 |
ENSMUST00000015576.6
|
Mcpt2
|
mast cell protease 2
|
|
chr8_+_4184652
Show fit
|
0.00 |
ENSMUST00000130372.3
|
Cd209g
|
CD209g antigen
|
|
chr10_+_25308466
Show fit
|
0.00 |
ENSMUST00000219224.2
ENSMUST00000219166.2
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2
|
|
chr6_+_146944253
Show fit
|
0.00 |
ENSMUST00000036111.10
|
Mrps35
|
mitochondrial ribosomal protein S35
|
|
chr3_+_98187743
Show fit
|
0.00 |
ENSMUST00000120541.8
ENSMUST00000090746.3
|
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2
|
|
chr16_-_22630327
Show fit
|
0.00 |
ENSMUST00000040592.6
|
Crygs
|
crystallin, gamma S
|
|
chr3_+_118355778
Show fit
|
0.00 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase
|
|
chr5_-_139799953
Show fit
|
0.00 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a
|
|
chr1_-_179572765
Show fit
|
0.00 |
ENSMUST00000211943.3
ENSMUST00000131716.4
ENSMUST00000221136.2
|
Kif28
|
kinesin family member 28
|
|
chr6_-_70364222
Show fit
|
0.00 |
ENSMUST00000103392.3
ENSMUST00000195945.2
|
Igkv8-16
|
immunoglobulin kappa variable 8-16
|
|
chr7_-_28329924
Show fit
|
0.00 |
ENSMUST00000159095.2
ENSMUST00000159418.8
ENSMUST00000159560.3
|
Acp7
|
acid phosphatase 7, tartrate resistant
|
|
chr9_-_38934656
Show fit
|
0.00 |
ENSMUST00000215383.2
ENSMUST00000214410.2
ENSMUST00000214369.2
|
Olfr146
|
olfactory receptor 146
|
|
chr5_-_26193648
Show fit
|
0.00 |
ENSMUST00000191203.7
|
Gm21698
|
predicted gene, 21698
|
|
chr5_-_26227914
Show fit
|
0.00 |
ENSMUST00000072286.7
|
Gm5862
|
predicted gene 5862
|
|
chr18_-_44292952
Show fit
|
0.00 |
ENSMUST00000181652.9
|
Gm10267
|
predicted gene 10267
|
|
chr18_+_13139992
Show fit
|
0.00 |
ENSMUST00000041676.3
ENSMUST00000234084.2
ENSMUST00000234565.2
|
Hrh4
|
histamine receptor H4
|
|
chr6_+_70680515
Show fit
|
0.00 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1
|
|
chr8_-_34333573
Show fit
|
0.00 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing
|