Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
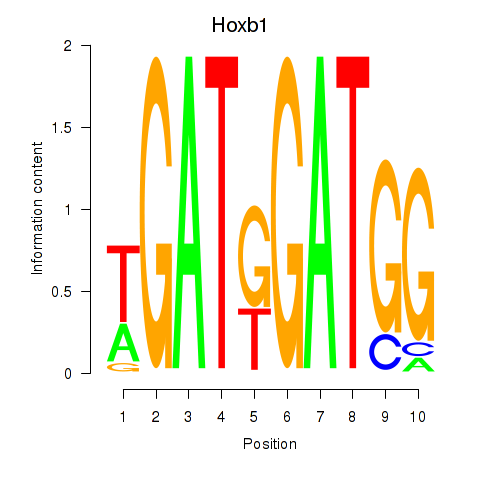
Results for Hoxb1
Z-value: 0.75
Transcription factors associated with Hoxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb1
|
ENSMUSG00000018973.3 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb1 | mm39_v1_chr11_+_96256565_96256578 | 0.96 | 1.0e-02 | Click! |
Activity profile of Hoxb1 motif
Sorted Z-values of Hoxb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127512933 | 0.60 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr11_-_97913420 | 0.54 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr8_-_106198112 | 0.36 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chrX_+_109857866 | 0.35 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr10_-_12745109 | 0.33 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr14_-_69740670 | 0.29 |
ENSMUST00000180059.3
ENSMUST00000179116.3 |
Gm16867
|
predicted gene, 16867 |
| chr14_-_69522431 | 0.29 |
ENSMUST00000183882.2
ENSMUST00000037064.5 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr10_+_4216353 | 0.28 |
ENSMUST00000045730.7
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr2_+_124452194 | 0.28 |
ENSMUST00000051419.15
ENSMUST00000076335.12 ENSMUST00000078621.12 ENSMUST00000077847.12 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr8_+_108669276 | 0.27 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr8_+_121262528 | 0.23 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr14_+_50656083 | 0.22 |
ENSMUST00000216949.2
|
Olfr739
|
olfactory receptor 739 |
| chr4_+_102971581 | 0.21 |
ENSMUST00000106858.8
|
Mier1
|
MEIR1 treanscription regulator |
| chr11_+_96256565 | 0.21 |
ENSMUST00000019117.3
|
Hoxb1
|
homeobox B1 |
| chr19_-_5553804 | 0.21 |
ENSMUST00000189704.2
|
Nscme3l
|
NSE3 homolog, SMC5-SMC6 complex component like |
| chr12_+_38830081 | 0.20 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr19_-_47452557 | 0.19 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr2_-_104324035 | 0.18 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr9_-_91247809 | 0.17 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr18_+_42644552 | 0.15 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr3_-_37286714 | 0.15 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr2_+_119803230 | 0.15 |
ENSMUST00000229024.2
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chrX_+_165127688 | 0.15 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr4_+_100633860 | 0.14 |
ENSMUST00000030257.15
ENSMUST00000097955.3 |
Cachd1
|
cache domain containing 1 |
| chr5_-_107437427 | 0.13 |
ENSMUST00000031224.15
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr5_-_138270995 | 0.13 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr10_+_67021509 | 0.13 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr4_+_43669266 | 0.12 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr2_+_84670956 | 0.12 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr9_+_78303059 | 0.11 |
ENSMUST00000113367.2
|
Ddx43
|
DEAD box helicase 43 |
| chr11_+_85243970 | 0.11 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr6_-_136852792 | 0.10 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr4_-_102971752 | 0.10 |
ENSMUST00000106868.4
|
Dnai4
|
dynein axonemal intermediate chain 4 |
| chr18_-_56695333 | 0.10 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr6_+_142359099 | 0.09 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr2_-_153371861 | 0.09 |
ENSMUST00000035346.14
|
Nol4l
|
nucleolar protein 4-like |
| chr2_-_90900525 | 0.08 |
ENSMUST00000153367.2
ENSMUST00000079976.10 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr17_+_27160203 | 0.08 |
ENSMUST00000194598.6
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr2_-_165717697 | 0.08 |
ENSMUST00000153655.8
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chrX_-_7999009 | 0.08 |
ENSMUST00000130832.8
ENSMUST00000033506.13 ENSMUST00000115623.8 ENSMUST00000153839.2 |
Wdr13
|
WD repeat domain 13 |
| chr2_+_119803180 | 0.07 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr2_-_90900628 | 0.07 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr17_+_27160356 | 0.06 |
ENSMUST00000229490.2
ENSMUST00000201702.5 ENSMUST00000177932.7 ENSMUST00000201349.6 |
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr4_+_102971909 | 0.06 |
ENSMUST00000143417.8
|
Mier1
|
MEIR1 treanscription regulator |
| chr6_-_87510200 | 0.06 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr9_-_21223631 | 0.05 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr1_-_135615831 | 0.05 |
ENSMUST00000190298.8
|
Nav1
|
neuron navigator 1 |
| chr11_-_83189670 | 0.04 |
ENSMUST00000175741.8
ENSMUST00000176518.2 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr16_-_97763780 | 0.04 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr12_+_79255702 | 0.03 |
ENSMUST00000122227.8
|
Rdh12
|
retinol dehydrogenase 12 |
| chr12_+_79255678 | 0.03 |
ENSMUST00000021548.12
|
Rdh12
|
retinol dehydrogenase 12 |
| chr1_+_153541020 | 0.03 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr19_-_3625698 | 0.02 |
ENSMUST00000172362.3
ENSMUST00000025846.16 ENSMUST00000226109.2 ENSMUST00000113997.9 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr18_-_56695288 | 0.02 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr11_+_76563281 | 0.02 |
ENSMUST00000056184.2
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr1_-_9770434 | 0.02 |
ENSMUST00000088658.11
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr2_+_124452493 | 0.01 |
ENSMUST00000103239.10
ENSMUST00000103240.9 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr19_-_41732104 | 0.01 |
ENSMUST00000025993.10
|
Slit1
|
slit guidance ligand 1 |
| chr14_-_24054352 | 0.01 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_157240138 | 0.01 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:0060938 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |