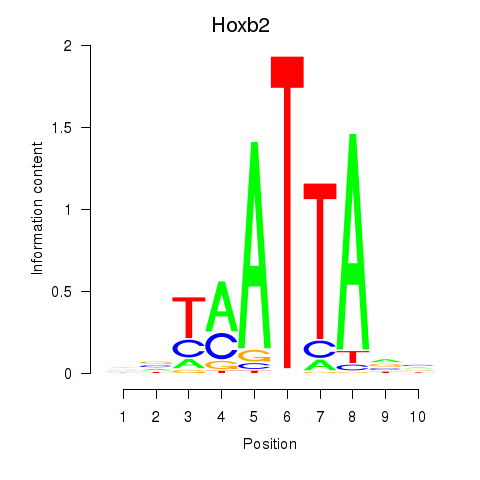
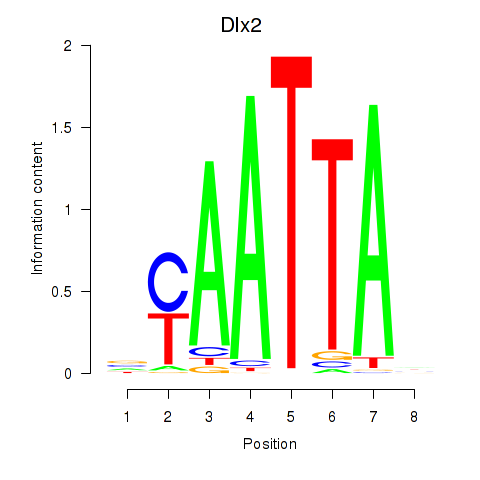
chr13_+_23728222
Show fit
1.93
ENSMUST00000075558.5
H3c7
H3 clustered histone 7
chr9_+_123195986
Show fit
1.86
ENSMUST00000038863.9
ENSMUST00000216843.2
Lars2
leucyl-tRNA synthetase, mitochondrial
chr2_+_87137925
Show fit
1.73
ENSMUST00000216396.3
Olfr1118
olfactory receptor 1118
chr5_-_84565218
Show fit
1.71
ENSMUST00000113401.4
Epha5
Eph receptor A5
chr5_-_62923463
Show fit
1.47
ENSMUST00000076623.8
ENSMUST00000159470.3
Arap2
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
chr2_-_111843053
Show fit
1.35
ENSMUST00000213559.3
Olfr1310
olfactory receptor 1310
chr2_-_45000389
Show fit
1.30
ENSMUST00000201804.4
ENSMUST00000028229.13
ENSMUST00000202187.4
ENSMUST00000153561.6
ENSMUST00000201490.2
Zeb2
zinc finger E-box binding homeobox 2
chr16_-_48592372
Show fit
1.21
ENSMUST00000231701.3
Trat1
T cell receptor associated transmembrane adaptor 1
chr10_-_128918779
Show fit
1.12
ENSMUST00000213579.2
Olfr767
olfactory receptor 767
chr2_+_83554770
Show fit
1.09
ENSMUST00000141725.3
Itgav
integrin alpha V
chr7_+_100435548
Show fit
1.06
ENSMUST00000216021.2
Fam168a
family with sequence similarity 168, member A
chr4_+_8690398
Show fit
1.04
ENSMUST00000127476.8
Chd7
chromodomain helicase DNA binding protein 7
chrM_+_9870
Show fit
1.03
ENSMUST00000084013.1
mt-Nd4l
mitochondrially encoded NADH dehydrogenase 4L
chr15_+_31224555
Show fit
1.01
ENSMUST00000186109.2
Dap
death-associated protein
chr19_-_13126896
Show fit
0.94
ENSMUST00000213493.2
Olfr1459
olfactory receptor 1459
chr6_-_116693849
Show fit
0.89
ENSMUST00000056623.13
Tmem72
transmembrane protein 72
chr11_-_73382303
Show fit
0.87
ENSMUST00000119863.2
ENSMUST00000215358.2
ENSMUST00000214623.2
Olfr381
olfactory receptor 381
chrM_+_11735
Show fit
0.85
ENSMUST00000082418.1
mt-Nd5
mitochondrially encoded NADH dehydrogenase 5
chr7_-_102998876
Show fit
0.82
ENSMUST00000215042.2
Olfr600
olfactory receptor 600
chr2_-_111880531
Show fit
0.80
ENSMUST00000213582.2
ENSMUST00000213961.3
ENSMUST00000215531.2
Olfr1312
olfactory receptor 1312
chr13_+_22508759
Show fit
0.80
ENSMUST00000226225.2
ENSMUST00000227017.2
Vmn1r197
vomeronasal 1 receptor 197
chr2_-_45001141
Show fit
0.79
ENSMUST00000201969.4
ENSMUST00000201623.4
Zeb2
zinc finger E-box binding homeobox 2
chr9_+_20193647
Show fit
0.77
ENSMUST00000071725.4
ENSMUST00000212983.3
Olfr39
olfactory receptor 39
chr17_-_37523969
Show fit
0.76
ENSMUST00000060728.7
ENSMUST00000216318.2
Olfr95
olfactory receptor 95
chr7_+_140181182
Show fit
0.76
ENSMUST00000214180.2
ENSMUST00000211771.2
Olfr46
olfactory receptor 46
chr17_+_21031817
Show fit
0.75
ENSMUST00000232810.2
ENSMUST00000233712.2
ENSMUST00000232852.2
Vmn1r229
vomeronasal 1 receptor 229
chr15_+_31224616
Show fit
0.74
ENSMUST00000186547.7
Dap
death-associated protein
chr13_-_23302027
Show fit
0.73
ENSMUST00000228656.2
Vmn1r217
vomeronasal 1 receptor 217
chr15_-_66985760
Show fit
0.73
ENSMUST00000092640.6
St3gal1
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
chr5_+_107655487
Show fit
0.72
ENSMUST00000143074.2
Gm42669
predicted gene 42669
chr1_-_140111138
Show fit
0.72
ENSMUST00000111976.9
ENSMUST00000066859.13
Cfh
complement component factor h
chr13_-_23302396
Show fit
0.70
ENSMUST00000227110.2
Vmn1r217
vomeronasal 1 receptor 217
chr8_-_69541852
Show fit
0.68
ENSMUST00000037478.13
ENSMUST00000148856.2
Slc18a1
solute carrier family 18 (vesicular monoamine), member 1
chr16_-_58620631
Show fit
0.66
ENSMUST00000206205.3
Olfr173
olfactory receptor 173
chr17_-_29226886
Show fit
0.65
ENSMUST00000232723.2
Stk38
serine/threonine kinase 38
chr17_-_37974666
Show fit
0.65
ENSMUST00000215414.2
ENSMUST00000213638.3
Olfr117
olfactory receptor 117
chr9_+_19716202
Show fit
0.64
ENSMUST00000212540.3
ENSMUST00000217280.2
Olfr859
olfactory receptor 859
chr1_+_151446893
Show fit
0.64
ENSMUST00000134499.8
Niban1
niban apoptosis regulator 1
chr10_-_85847697
Show fit
0.63
ENSMUST00000105304.2
ENSMUST00000061699.12
Bpifc
BPI fold containing family C
chr10_+_53213763
Show fit
0.62
ENSMUST00000219491.2
ENSMUST00000163319.9
ENSMUST00000220197.2
ENSMUST00000046221.8
ENSMUST00000218468.2
ENSMUST00000219921.2
Pln
phospholamban
chr3_-_129834788
Show fit
0.62
ENSMUST00000168644.3
Sec24b
Sec24 related gene family, member B (S. cerevisiae)
chr5_-_116162415
Show fit
0.61
ENSMUST00000031486.14
ENSMUST00000111999.8
Prkab1
protein kinase, AMP-activated, beta 1 non-catalytic subunit
chr13_-_59970383
Show fit
0.60
ENSMUST00000225987.2
Tut7
terminal uridylyl transferase 7
chr7_+_23400128
Show fit
0.59
ENSMUST00000226233.2
ENSMUST00000227987.2
Vmn1r173
vomeronasal 1 receptor 173
chr19_-_12313274
Show fit
0.59
ENSMUST00000208398.3
Olfr1438-ps1
olfactory receptor 1438, pseudogene 1
chr2_-_111820618
Show fit
0.58
ENSMUST00000216948.2
ENSMUST00000214935.2
ENSMUST00000217452.2
ENSMUST00000215045.2
Olfr1309
olfactory receptor 1309
chr2_-_45000250
Show fit
0.56
ENSMUST00000201211.4
ENSMUST00000177302.8
Zeb2
zinc finger E-box binding homeobox 2
chr3_+_60380243
Show fit
0.55
ENSMUST00000195724.6
Mbnl1
muscleblind like splicing factor 1
chr9_+_39932760
Show fit
0.55
ENSMUST00000215956.3
Olfr981
olfactory receptor 981
chr12_-_99529767
Show fit
0.55
ENSMUST00000176928.3
ENSMUST00000223484.2
Foxn3
forkhead box N3
chr8_-_3675525
Show fit
0.55
ENSMUST00000144977.2
ENSMUST00000136105.8
ENSMUST00000128566.8
Pcp2
Purkinje cell protein 2 (L7)
chr1_+_173983199
Show fit
0.55
ENSMUST00000213748.2
Olfr420
olfactory receptor 420
chr3_-_15902583
Show fit
0.54
ENSMUST00000108354.8
ENSMUST00000108349.2
ENSMUST00000108352.9
ENSMUST00000108350.8
ENSMUST00000050623.11
Sirpb1c
signal-regulatory protein beta 1C
chr10_-_44024843
Show fit
0.54
ENSMUST00000200401.2
Crybg1
crystallin beta-gamma domain containing 1
chr2_+_129854256
Show fit
0.52
ENSMUST00000110299.3
Tgm3
transglutaminase 3, E polypeptide
chr13_+_23317325
Show fit
0.52
ENSMUST00000227050.2
ENSMUST00000227160.2
ENSMUST00000227741.2
ENSMUST00000226692.2
Vmn1r218
vomeronasal 1 receptor 218
chrX_-_142716200
Show fit
0.52
ENSMUST00000112851.8
ENSMUST00000112856.3
ENSMUST00000033642.10
Dcx
doublecortin
chr7_+_103199575
Show fit
0.52
ENSMUST00000106888.3
Olfr613
olfactory receptor 613
chr8_+_4288815
Show fit
0.52
ENSMUST00000003027.14
ENSMUST00000110999.8
Map2k7
mitogen-activated protein kinase kinase 7
chr1_-_92412835
Show fit
0.51
ENSMUST00000214928.3
Olfr1416
olfactory receptor 1416
chr4_+_108576846
Show fit
0.51
ENSMUST00000178992.2
3110021N24Rik
RIKEN cDNA 3110021N24 gene
chr4_+_19818718
Show fit
0.50
ENSMUST00000035890.8
Slc7a13
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13
chr1_-_140111018
Show fit
0.50
ENSMUST00000192880.6
ENSMUST00000111977.8
Cfh
complement component factor h
chr1_+_107456731
Show fit
0.49
ENSMUST00000182198.8
Serpinb10
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10
chr5_-_116427003
Show fit
0.49
ENSMUST00000086483.4
ENSMUST00000050178.13
Ccdc60
coiled-coil domain containing 60
chr16_+_33504740
Show fit
0.48
ENSMUST00000232568.2
Heg1
heart development protein with EGF-like domains 1
chr6_+_136509922
Show fit
0.47
ENSMUST00000187429.4
Atf7ip
activating transcription factor 7 interacting protein
chr2_+_152596075
Show fit
0.47
ENSMUST00000010020.12
Cox4i2
cytochrome c oxidase subunit 4I2
chr12_+_38830081
Show fit
0.47
ENSMUST00000095767.11
Etv1
ets variant 1
chr13_+_23281146
Show fit
0.47
ENSMUST00000228389.2
Vmn1r216
vomeronasal 1 receptor 216
chr2_-_88157559
Show fit
0.46
ENSMUST00000214207.2
Olfr1175
olfactory receptor 1175
chr13_+_23214588
Show fit
0.46
ENSMUST00000227652.2
ENSMUST00000227236.2
Vmn1r214
vomeronasal 1 receptor 214
chr2_-_111779785
Show fit
0.45
ENSMUST00000099604.6
Olfr1307
olfactory receptor 1307
chr10_+_127256736
Show fit
0.45
ENSMUST00000064793.13
R3hdm2
R3H domain containing 2
chr3_+_90201388
Show fit
0.45
ENSMUST00000199607.5
Gatad2b
GATA zinc finger domain containing 2B
chr3_+_63988968
Show fit
0.44
ENSMUST00000029406.6
Vmn2r1
vomeronasal 2, receptor 1
chr7_-_103734672
Show fit
0.44
ENSMUST00000057104.7
Olfr645
olfactory receptor 645
chr8_-_5155347
Show fit
0.44
ENSMUST00000023835.3
Slc10a2
solute carrier family 10, member 2
chr11_-_50183129
Show fit
0.44
ENSMUST00000059458.5
Maml1
mastermind like transcriptional coactivator 1
chrX_-_156198282
Show fit
0.44
ENSMUST00000079945.11
ENSMUST00000138396.3
Phex
phosphate regulating endopeptidase homolog, X-linked
chr6_-_57992144
Show fit
0.43
ENSMUST00000228070.2
ENSMUST00000228040.2
Vmn1r26
vomeronasal 1 receptor 26
chrX_+_158491589
Show fit
0.43
ENSMUST00000080394.13
Sh3kbp1
SH3-domain kinase binding protein 1
chr12_-_111780268
Show fit
0.43
ENSMUST00000021715.6
Xrcc3
X-ray repair complementing defective repair in Chinese hamster cells 3
chr2_-_5680801
Show fit
0.42
ENSMUST00000114987.4
Camk1d
calcium/calmodulin-dependent protein kinase ID
chr17_+_20950105
Show fit
0.42
ENSMUST00000233597.2
ENSMUST00000232943.2
ENSMUST00000179914.3
Vmn1r227
vomeronasal 1 receptor 227
chr1_-_155293141
Show fit
0.42
ENSMUST00000111775.8
ENSMUST00000111774.2
Xpr1
xenotropic and polytropic retrovirus receptor 1
chr11_-_96859484
Show fit
0.42
ENSMUST00000107623.8
Sp2
Sp2 transcription factor
chr1_+_104696235
Show fit
0.41
ENSMUST00000062528.9
Cdh20
cadherin 20
chr2_-_104324035
Show fit
0.41
ENSMUST00000111124.8
Hipk3
homeodomain interacting protein kinase 3
chr12_+_76580386
Show fit
0.41
ENSMUST00000219063.2
Plekhg3
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
chr7_-_12829100
Show fit
0.40
ENSMUST00000209822.3
ENSMUST00000235753.2
Vmn1r85
vomeronasal 1 receptor 85
chr9_+_39556381
Show fit
0.40
ENSMUST00000219295.2
Olfr961
olfactory receptor 961
chr9_+_40092216
Show fit
0.40
ENSMUST00000218134.2
ENSMUST00000216720.2
ENSMUST00000214763.2
Olfr986
olfactory receptor 986
chr13_+_22656093
Show fit
0.40
ENSMUST00000226330.2
ENSMUST00000226965.2
Vmn1r201
vomeronasal 1 receptor 201
chr10_+_78816884
Show fit
0.40
ENSMUST00000058991.5
ENSMUST00000203973.2
Olfr1352
olfactory receptor 1352
chr11_-_73348284
Show fit
0.40
ENSMUST00000121209.3
ENSMUST00000127789.3
Olfr380
olfactory receptor 380
chr7_-_103535459
Show fit
0.40
ENSMUST00000216303.2
Olfr66
olfactory receptor 66
chr7_+_3648264
Show fit
0.39
ENSMUST00000206287.2
ENSMUST00000038913.16
Cnot3
CCR4-NOT transcription complex, subunit 3
chr2_-_165997551
Show fit
0.39
ENSMUST00000109249.9
ENSMUST00000146497.9
Sulf2
sulfatase 2
chr2_-_45007407
Show fit
0.39
ENSMUST00000176438.9
Zeb2
zinc finger E-box binding homeobox 2
chr4_-_15149755
Show fit
0.39
ENSMUST00000108273.2
Necab1
N-terminal EF-hand calcium binding protein 1
chrM_+_7006
Show fit
0.38
ENSMUST00000082405.1
mt-Co2
mitochondrially encoded cytochrome c oxidase II
chr14_-_50586329
Show fit
0.38
ENSMUST00000216634.2
Olfr735
olfactory receptor 735
chr17_-_21216726
Show fit
0.38
ENSMUST00000237195.2
ENSMUST00000237629.2
ENSMUST00000056339.3
Vmn1r233
vomeronasal 1 receptor 233
chr15_-_71826243
Show fit
0.38
ENSMUST00000229585.2
Col22a1
collagen, type XXII, alpha 1
chr2_-_45002902
Show fit
0.38
ENSMUST00000076836.13
ENSMUST00000176732.8
ENSMUST00000200844.4
Zeb2
zinc finger E-box binding homeobox 2
chr8_-_32440071
Show fit
0.38
ENSMUST00000207678.3
Nrg1
neuregulin 1
chrM_+_2743
Show fit
0.37
ENSMUST00000082392.1
mt-Nd1
mitochondrially encoded NADH dehydrogenase 1
chr2_-_44912927
Show fit
0.37
ENSMUST00000202432.4
Zeb2
zinc finger E-box binding homeobox 2
chr7_+_106737534
Show fit
0.37
ENSMUST00000213367.3
ENSMUST00000214819.3
ENSMUST00000216871.3
ENSMUST00000215284.3
ENSMUST00000209942.2
Olfr716
olfactory receptor 716
chr7_-_10292412
Show fit
0.37
ENSMUST00000236246.2
Vmn1r68
vomeronasal 1 receptor 68
chrX_+_158623460
Show fit
0.37
ENSMUST00000112451.8
ENSMUST00000112453.9
Sh3kbp1
SH3-domain kinase binding protein 1
chr2_+_37101586
Show fit
0.37
ENSMUST00000214897.2
Olfr366
olfactory receptor 366
chr13_+_23758555
Show fit
0.37
ENSMUST00000090776.7
H2ac7
H2A clustered histone 7
chr10_-_12689345
Show fit
0.37
ENSMUST00000217899.2
Utrn
utrophin
chr13_-_53627110
Show fit
0.36
ENSMUST00000021922.10
Msx2
msh homeobox 2
chr17_-_29226700
Show fit
0.35
ENSMUST00000233441.2
Stk38
serine/threonine kinase 38
chr7_+_63835285
Show fit
0.35
ENSMUST00000206263.2
ENSMUST00000206107.2
ENSMUST00000205731.2
ENSMUST00000206706.2
ENSMUST00000205690.2
Trpm1
transient receptor potential cation channel, subfamily M, member 1
chr12_-_31763859
Show fit
0.35
ENSMUST00000057783.6
ENSMUST00000236002.2
ENSMUST00000174480.3
ENSMUST00000176710.2
Gpr22
G protein-coupled receptor 22
chr14_+_47536075
Show fit
0.34
ENSMUST00000227554.2
Mapk1ip1l
mitogen-activated protein kinase 1 interacting protein 1-like
chrM_+_10167
Show fit
0.34
ENSMUST00000082414.1
mt-Nd4
mitochondrially encoded NADH dehydrogenase 4
chr7_+_107585900
Show fit
0.33
ENSMUST00000214677.2
Olfr477
olfactory receptor 477
chr14_+_8283087
Show fit
0.33
ENSMUST00000206298.3
ENSMUST00000216079.2
Olfr720
olfactory receptor 720
chr13_+_23191826
Show fit
0.33
ENSMUST00000228758.2
ENSMUST00000228031.2
ENSMUST00000227573.2
Vmn1r213
vomeronasal 1 receptor 213
chr14_-_8146867
Show fit
0.33
ENSMUST00000217035.2
ENSMUST00000206009.3
Olfr31
olfactory receptor 31
chr17_-_37472385
Show fit
0.33
ENSMUST00000219235.3
Olfr93
olfactory receptor 93
chr2_-_153079828
Show fit
0.32
ENSMUST00000109795.2
Plagl2
pleiomorphic adenoma gene-like 2
chr7_-_28246530
Show fit
0.32
ENSMUST00000239002.2
ENSMUST00000057974.4
Nccrp1
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish)
chrX_+_164953444
Show fit
0.32
ENSMUST00000130880.9
ENSMUST00000056410.11
ENSMUST00000096252.10
ENSMUST00000087169.11
Gemin8
gem nuclear organelle associated protein 8
chr5_+_42225303
Show fit
0.32
ENSMUST00000087332.5
Gm16223
predicted gene 16223
chr8_+_4288733
Show fit
0.32
ENSMUST00000110998.9
ENSMUST00000062686.11
Map2k7
mitogen-activated protein kinase kinase 7
chr5_-_86521273
Show fit
0.32
ENSMUST00000031175.12
Tmprss11d
transmembrane protease, serine 11d
chr17_+_12338161
Show fit
0.31
ENSMUST00000024594.9
Agpat4
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
chr2_-_165997179
Show fit
0.31
ENSMUST00000088086.4
Sulf2
sulfatase 2
chr8_-_26609153
Show fit
0.31
ENSMUST00000037182.14
Hook3
hook microtubule tethering protein 3
chr18_+_4920513
Show fit
0.31
ENSMUST00000126977.8
Svil
supervillin
chr19_+_13208692
Show fit
0.31
ENSMUST00000207246.4
Olfr1463
olfactory receptor 1463
chr10_-_30647836
Show fit
0.31
ENSMUST00000215926.2
ENSMUST00000213836.2
Ncoa7
nuclear receptor coactivator 7
chr7_-_100613579
Show fit
0.31
ENSMUST00000060174.6
P2ry6
pyrimidinergic receptor P2Y, G-protein coupled, 6
chr11_-_73742280
Show fit
0.30
ENSMUST00000213365.2
Olfr393
olfactory receptor 393
chr8_-_32408864
Show fit
0.30
ENSMUST00000073884.7
ENSMUST00000238812.2
Nrg1
neuregulin 1
chr17_-_29226965
Show fit
0.30
ENSMUST00000009138.13
ENSMUST00000119274.3
Stk38
serine/threonine kinase 38
chr6_-_97436223
Show fit
0.29
ENSMUST00000113359.8
Frmd4b
FERM domain containing 4B
chr5_+_4073343
Show fit
0.28
ENSMUST00000238634.2
Akap9
A kinase (PRKA) anchor protein (yotiao) 9
chr13_-_23372145
Show fit
0.28
ENSMUST00000228239.2
ENSMUST00000227950.2
ENSMUST00000226651.2
ENSMUST00000227679.2
ENSMUST00000228854.2
Vmn1r220
vomeronasal 1 receptor 220
chr13_+_75855695
Show fit
0.28
ENSMUST00000222194.2
ENSMUST00000223535.2
ENSMUST00000222853.2
Ell2
elongation factor for RNA polymerase II 2
chr11_+_29323618
Show fit
0.28
ENSMUST00000040182.13
ENSMUST00000109477.2
Ccdc88a
coiled coil domain containing 88A
chr10_+_73657689
Show fit
0.28
ENSMUST00000064562.14
ENSMUST00000193174.6
ENSMUST00000105426.10
ENSMUST00000129404.9
ENSMUST00000131321.9
ENSMUST00000126920.9
ENSMUST00000147189.9
ENSMUST00000105424.10
ENSMUST00000092420.13
ENSMUST00000105429.10
ENSMUST00000193361.6
ENSMUST00000131724.9
ENSMUST00000152655.9
ENSMUST00000151116.9
ENSMUST00000155701.9
ENSMUST00000152819.9
ENSMUST00000125517.9
ENSMUST00000124046.8
ENSMUST00000146682.8
ENSMUST00000177107.8
ENSMUST00000149977.9
ENSMUST00000191854.6
Pcdh15
protocadherin 15
chr13_+_83723743
Show fit
0.27
ENSMUST00000198217.5
ENSMUST00000199210.5
Mef2c
myocyte enhancer factor 2C
chr5_-_23881353
Show fit
0.27
ENSMUST00000198661.5
Srpk2
serine/arginine-rich protein specific kinase 2
chr3_+_60380463
Show fit
0.27
ENSMUST00000195077.6
ENSMUST00000193647.6
ENSMUST00000195001.2
ENSMUST00000192807.6
Mbnl1
muscleblind like splicing factor 1
chr16_+_42727926
Show fit
0.27
ENSMUST00000151244.8
ENSMUST00000114694.9
Zbtb20
zinc finger and BTB domain containing 20
chr19_-_12100560
Show fit
0.27
ENSMUST00000213471.2
ENSMUST00000214676.2
Olfr76
olfactory receptor 76
chr7_+_126184108
Show fit
0.27
ENSMUST00000039522.8
Apobr
apolipoprotein B receptor
chr13_+_118851214
Show fit
0.26
ENSMUST00000022246.9
Fgf10
fibroblast growth factor 10
chr14_-_52277310
Show fit
0.26
ENSMUST00000216907.2
ENSMUST00000214071.2
ENSMUST00000216188.2
Olfr221
olfactory receptor 221
chr7_+_23451695
Show fit
0.26
ENSMUST00000228331.2
Vmn1r174
vomeronasal 1 receptor 174
chr6_+_90078412
Show fit
0.26
ENSMUST00000089417.8
ENSMUST00000226577.2
Vmn1r50
vomeronasal 1 receptor 50
chr2_-_32976378
Show fit
0.25
ENSMUST00000049618.9
Garnl3
GTPase activating RANGAP domain-like 3
chr7_+_84502761
Show fit
0.25
ENSMUST00000217039.3
ENSMUST00000211582.2
Olfr291
olfactory receptor 291
chr15_-_43733389
Show fit
0.25
ENSMUST00000067469.6
Tmem74
transmembrane protein 74
chr17_+_21446349
Show fit
0.25
ENSMUST00000235895.2
Vmn1r234
vomeronasal 1 receptor 234
chr16_+_44914397
Show fit
0.24
ENSMUST00000061050.6
Ccdc80
coiled-coil domain containing 80
chr2_-_37007795
Show fit
0.24
ENSMUST00000213817.2
ENSMUST00000215927.2
Olfr362
olfactory receptor 362
chr10_-_30647881
Show fit
0.24
ENSMUST00000215740.2
Ncoa7
nuclear receptor coactivator 7
chr18_+_82932747
Show fit
0.24
ENSMUST00000071233.7
Zfp516
zinc finger protein 516
chr2_+_22512195
Show fit
0.24
ENSMUST00000028123.4
Gad2
glutamic acid decarboxylase 2
chr1_+_92545510
Show fit
0.24
ENSMUST00000213247.2
Olfr12
olfactory receptor 12
chr3_-_146388165
Show fit
0.23
ENSMUST00000124931.8
ENSMUST00000147113.2
Samd13
sterile alpha motif domain containing 13
chr2_+_111984716
Show fit
0.22
ENSMUST00000214063.2
ENSMUST00000217533.2
Olfr1318
olfactory receptor 1318
chr15_+_31225302
Show fit
0.22
ENSMUST00000186425.7
Dap
death-associated protein
chrX_+_23559282
Show fit
0.22
ENSMUST00000035766.13
ENSMUST00000101670.3
Wdr44
WD repeat domain 44
chr15_+_39522905
Show fit
0.21
ENSMUST00000226410.2
Rims2
regulating synaptic membrane exocytosis 2
chr8_+_23901506
Show fit
0.21
ENSMUST00000033952.8
Sfrp1
secreted frizzled-related protein 1
chr13_+_22534534
Show fit
0.21
ENSMUST00000226909.2
ENSMUST00000227167.2
ENSMUST00000226786.2
Vmn1r198
vomeronasal 1 receptor 198
chr18_+_32200781
Show fit
0.20
ENSMUST00000025243.5
ENSMUST00000212675.2
Iws1
IWS1, SUPT6 interacting protein
chr3_-_88368489
Show fit
0.20
ENSMUST00000166237.8
Sema4a
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
chr11_-_69556904
Show fit
0.20
ENSMUST00000018918.12
Cd68
CD68 antigen
chr8_-_32408380
Show fit
0.20
ENSMUST00000208497.3
ENSMUST00000207584.3
Nrg1
neuregulin 1
chr17_+_38456172
Show fit
0.19
ENSMUST00000215078.3
ENSMUST00000215549.3
ENSMUST00000173610.2
Olfr133
olfactory receptor 133
chr6_-_116084810
Show fit
0.19
ENSMUST00000204353.3
Tmcc1
transmembrane and coiled coil domains 1
chr15_+_31224460
Show fit
0.19
ENSMUST00000044524.16
Dap
death-associated protein
chr7_+_126376099
Show fit
0.19
ENSMUST00000038614.12
ENSMUST00000170882.8
ENSMUST00000106359.2
ENSMUST00000106357.8
ENSMUST00000145762.8
Ypel3
yippee like 3
chr3_-_133250889
Show fit
0.19
ENSMUST00000197118.5
Tet2
tet methylcytosine dioxygenase 2
chr14_+_51333816
Show fit
0.19
ENSMUST00000169895.3
Rnase4
ribonuclease, RNase A family 4
chr19_+_12364643
Show fit
0.19
ENSMUST00000217062.3
ENSMUST00000216145.2
ENSMUST00000213657.2
Olfr1440
olfactory receptor 1440
chr10_+_127256993
Show fit
0.19
ENSMUST00000170336.8
R3hdm2
R3H domain containing 2
chr7_-_107726656
Show fit
0.18
ENSMUST00000214722.2
Olfr484
olfactory receptor 484
chr14_+_8282925
Show fit
0.18
ENSMUST00000217642.2
Olfr720
olfactory receptor 720
chr9_-_37627519
Show fit
0.18
ENSMUST00000215727.2
ENSMUST00000211952.3
Olfr160
olfactory receptor 160
chr7_-_84339156
Show fit
0.18
ENSMUST00000209117.2
ENSMUST00000207975.2
Zfand6
Links
zinc finger, AN1-type domain 6
chr2_-_87543523
Show fit
0.18
ENSMUST00000214209.2
Olfr1137
olfactory receptor 1137
chr17_-_37399343
Show fit
0.18
ENSMUST00000207101.2
ENSMUST00000217397.2
ENSMUST00000215195.2
ENSMUST00000216488.2
Olfr90
olfactory receptor 90
chr15_-_83033471
Show fit
0.18
ENSMUST00000129372.2
Poldip3
polymerase (DNA-directed), delta interacting protein 3
chr5_+_138083345
Show fit
0.18
ENSMUST00000019660.11
ENSMUST00000066617.12
ENSMUST00000110963.8
Zkscan1
zinc finger with KRAB and SCAN domains 1
chr3_+_94745009
Show fit
0.17
ENSMUST00000107266.8
ENSMUST00000042402.12
ENSMUST00000107269.2
Pogz
pogo transposable element with ZNF domain
chr12_-_56581823
Show fit
0.17
ENSMUST00000178477.9
Nkx2-1
NK2 homeobox 1
chr9_+_38725910
Show fit
0.17
ENSMUST00000213164.2
Olfr922
olfactory receptor 922
chr7_+_122723365
Show fit
0.17
ENSMUST00000205514.2
ENSMUST00000094053.7
Tnrc6a
trinucleotide repeat containing 6a
chr13_+_81034214
Show fit
0.17
ENSMUST00000161441.2
Arrdc3
arrestin domain containing 3