Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxb3
Z-value: 1.64
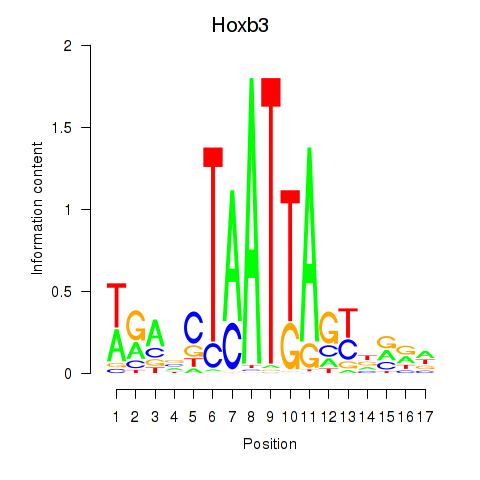
Transcription factors associated with Hoxb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb3
|
ENSMUSG00000048763.12 | homeobox B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb3 | mm39_v1_chr11_+_96214078_96214152 | -0.66 | 2.3e-01 | Click! |
Activity profile of Hoxb3 motif
Sorted Z-values of Hoxb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23929490 | 3.18 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr13_-_21900313 | 2.70 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr1_-_92412835 | 1.55 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr5_-_84565218 | 1.45 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr12_-_99529767 | 1.45 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr3_-_15902583 | 1.40 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr10_+_129539079 | 1.25 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
| chr2_-_111820618 | 1.10 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr4_-_155729865 | 1.10 |
ENSMUST00000115821.3
|
Gm10563
|
predicted gene 10563 |
| chr2_-_89423470 | 1.08 |
ENSMUST00000217254.2
ENSMUST00000217192.2 ENSMUST00000213221.2 |
Olfr1246
|
olfactory receptor 1246 |
| chr9_-_39618413 | 0.98 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149 |
| chr2_-_89951611 | 0.88 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr1_+_173876771 | 0.88 |
ENSMUST00000213381.2
ENSMUST00000213211.2 |
Olfr432
|
olfactory receptor 432 |
| chr10_-_128918779 | 0.87 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr7_+_3648264 | 0.84 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr16_-_58695131 | 0.79 |
ENSMUST00000217377.2
|
Olfr177
|
olfactory receptor 177 |
| chr4_-_131802606 | 0.77 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr5_-_137015683 | 0.74 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr7_-_10292412 | 0.74 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr7_+_23239157 | 0.72 |
ENSMUST00000235361.2
|
Vmn1r168
|
vomeronasal 1 receptor 168 |
| chr2_-_87868043 | 0.72 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr14_-_8146867 | 0.71 |
ENSMUST00000217035.2
ENSMUST00000206009.3 |
Olfr31
|
olfactory receptor 31 |
| chr2_-_111253457 | 0.71 |
ENSMUST00000213210.2
ENSMUST00000184954.4 |
Olfr1286
|
olfactory receptor 1286 |
| chr11_+_49410475 | 0.70 |
ENSMUST00000204706.3
|
Olfr1383
|
olfactory receptor 1383 |
| chr16_+_51851948 | 0.69 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_-_12829100 | 0.67 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr19_-_12302465 | 0.66 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chrM_+_9870 | 0.66 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr16_+_51851917 | 0.64 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr9_-_62895197 | 0.63 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chr7_+_28869770 | 0.62 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr2_-_111843053 | 0.62 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr9_-_119897358 | 0.58 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr14_-_70666513 | 0.58 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr14_+_99536111 | 0.56 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr6_+_90078412 | 0.56 |
ENSMUST00000089417.8
ENSMUST00000226577.2 |
Vmn1r50
|
vomeronasal 1 receptor 50 |
| chr2_-_111324108 | 0.53 |
ENSMUST00000208881.2
ENSMUST00000208695.2 ENSMUST00000217611.2 |
Olfr1290
|
olfactory receptor 1290 |
| chr1_-_150341911 | 0.52 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr9_-_119897328 | 0.52 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr5_+_63969706 | 0.51 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr2_-_5680801 | 0.51 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr3_-_15640045 | 0.50 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr10_-_128875765 | 0.49 |
ENSMUST00000204763.3
|
Olfr764-ps1
|
olfactory receptor 764, pseudogene 1 |
| chr4_-_41045381 | 0.49 |
ENSMUST00000054945.8
|
Aqp7
|
aquaporin 7 |
| chr12_-_25146078 | 0.48 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chr11_-_103247150 | 0.48 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr17_-_37399343 | 0.46 |
ENSMUST00000207101.2
ENSMUST00000217397.2 ENSMUST00000215195.2 ENSMUST00000216488.2 |
Olfr90
|
olfactory receptor 90 |
| chr11_-_95966477 | 0.45 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr19_+_12364643 | 0.45 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr1_-_92107971 | 0.44 |
ENSMUST00000186002.3
ENSMUST00000097644.9 |
Hdac4
|
histone deacetylase 4 |
| chr2_-_86257093 | 0.44 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr4_-_52859227 | 0.44 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr16_+_51851588 | 0.43 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr8_+_114362181 | 0.43 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr8_+_66838927 | 0.41 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr9_+_40092216 | 0.40 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr8_+_114362419 | 0.39 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr7_+_28869629 | 0.39 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr2_+_118877610 | 0.38 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr4_-_131802561 | 0.37 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr15_-_9529898 | 0.37 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr9_+_38725910 | 0.36 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chr7_+_4925781 | 0.35 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr2_-_86208737 | 0.33 |
ENSMUST00000217435.2
|
Olfr1057
|
olfactory receptor 1057 |
| chr2_-_32277773 | 0.32 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr2_-_90301592 | 0.32 |
ENSMUST00000111493.8
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr3_+_60503051 | 0.32 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr11_+_69471185 | 0.30 |
ENSMUST00000171247.8
ENSMUST00000108658.10 ENSMUST00000005371.12 |
Trp53
|
transformation related protein 53 |
| chr6_-_125357756 | 0.29 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr13_-_112698563 | 0.27 |
ENSMUST00000224510.2
|
Il31ra
|
interleukin 31 receptor A |
| chr10_-_75946790 | 0.27 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr2_+_3425159 | 0.26 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr7_-_44741622 | 0.26 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr4_-_3938352 | 0.26 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr7_-_86016045 | 0.26 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr8_+_23901506 | 0.25 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr11_-_73800125 | 0.25 |
ENSMUST00000215690.2
|
Olfr395
|
olfactory receptor 395 |
| chr4_-_114991478 | 0.25 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr12_+_112727089 | 0.25 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr13_-_74956640 | 0.25 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr18_+_61408073 | 0.25 |
ENSMUST00000135688.2
|
Pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr11_-_102076028 | 0.25 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr17_+_37148015 | 0.24 |
ENSMUST00000179968.8
ENSMUST00000130367.8 ENSMUST00000053434.15 ENSMUST00000130801.8 ENSMUST00000144182.8 ENSMUST00000123715.8 |
Trim26
|
tripartite motif-containing 26 |
| chr5_+_137015873 | 0.24 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr5_+_145217272 | 0.24 |
ENSMUST00000200246.2
|
Zscan25
|
zinc finger and SCAN domain containing 25 |
| chr1_-_92423800 | 0.23 |
ENSMUST00000204766.2
ENSMUST00000204009.3 |
Olfr1415
|
olfactory receptor 1415 |
| chr10_-_125164826 | 0.22 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr9_+_20209828 | 0.22 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr14_-_105414714 | 0.21 |
ENSMUST00000100327.10
ENSMUST00000022715.14 |
Rbm26
|
RNA binding motif protein 26 |
| chrX_-_49108236 | 0.21 |
ENSMUST00000213556.3
ENSMUST00000213463.2 |
Olfr1323
|
olfactory receptor 1323 |
| chr13_-_74956030 | 0.20 |
ENSMUST00000065629.6
|
Cast
|
calpastatin |
| chrY_+_5656986 | 0.20 |
ENSMUST00000190391.2
|
Gm21854
|
predicted gene, 21854 |
| chr7_+_6441687 | 0.20 |
ENSMUST00000218906.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr2_+_118877594 | 0.20 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr17_-_45970238 | 0.20 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr6_-_89825033 | 0.19 |
ENSMUST00000226436.2
|
Vmn1r42
|
vomeronasal 1 receptor 42 |
| chr11_+_5738480 | 0.19 |
ENSMUST00000109845.8
ENSMUST00000020769.14 ENSMUST00000102928.5 |
Dbnl
|
drebrin-like |
| chr13_-_53627110 | 0.19 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr18_+_36877709 | 0.19 |
ENSMUST00000007042.6
ENSMUST00000237095.2 |
Ik
|
IK cytokine |
| chr2_-_160950936 | 0.19 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr2_+_69727599 | 0.18 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_87854404 | 0.18 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chrY_+_5942460 | 0.17 |
ENSMUST00000185926.2
|
Gm21778
|
predicted gene, 21778 |
| chr14_+_50360643 | 0.17 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr6_-_89853395 | 0.16 |
ENSMUST00000227279.2
ENSMUST00000228709.2 ENSMUST00000226983.2 |
Vmn1r42
Vmn1r43
|
vomeronasal 1 receptor 42 vomeronasal 1 receptor 43 |
| chr11_+_121128042 | 0.16 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor |
| chr2_-_165230165 | 0.16 |
ENSMUST00000103084.4
|
Zfp334
|
zinc finger protein 334 |
| chr6_+_65019558 | 0.16 |
ENSMUST00000204801.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_+_23727325 | 0.16 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr10_-_83484467 | 0.15 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr2_+_32178325 | 0.15 |
ENSMUST00000100194.10
ENSMUST00000131712.9 ENSMUST00000081670.14 ENSMUST00000147707.8 ENSMUST00000129193.3 |
Golga2
|
golgi autoantigen, golgin subfamily a, 2 |
| chr2_+_87436556 | 0.14 |
ENSMUST00000213103.3
ENSMUST00000216580.2 |
Olfr1130
|
olfactory receptor 1130 |
| chr5_-_143279378 | 0.14 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr10_-_128885867 | 0.14 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr7_-_12103200 | 0.14 |
ENSMUST00000228653.2
ENSMUST00000227427.2 ENSMUST00000226408.2 |
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chr1_-_192880260 | 0.13 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr1_-_33853598 | 0.13 |
ENSMUST00000019861.13
ENSMUST00000044455.8 |
Zfp451
|
zinc finger protein 451 |
| chr2_-_25351106 | 0.12 |
ENSMUST00000114261.9
|
Paxx
|
non-homologous end joining factor |
| chr17_+_85335775 | 0.12 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr17_+_17622934 | 0.12 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr12_-_111150925 | 0.12 |
ENSMUST00000121608.2
|
4930595D18Rik
|
RIKEN cDNA 4930595D18 gene |
| chr13_+_46655324 | 0.12 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chrM_+_10167 | 0.11 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr12_-_114672701 | 0.11 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr3_-_15491482 | 0.10 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr19_-_5610628 | 0.09 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr9_+_35819708 | 0.09 |
ENSMUST00000176049.2
ENSMUST00000176153.2 |
Pate13
|
prostate and testis expressed 13 |
| chr5_-_137784912 | 0.08 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr1_+_40363701 | 0.08 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr17_-_57338468 | 0.08 |
ENSMUST00000007814.10
ENSMUST00000233480.2 |
Khsrp
|
KH-type splicing regulatory protein |
| chr2_-_132089667 | 0.08 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr16_-_30086317 | 0.08 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr6_-_70383976 | 0.07 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr6_-_122317484 | 0.07 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr4_-_43710231 | 0.07 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr16_-_19341016 | 0.06 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr6_-_41752111 | 0.06 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr15_+_10952418 | 0.06 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr18_+_12874390 | 0.06 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr17_-_45785752 | 0.05 |
ENSMUST00000233553.2
ENSMUST00000233769.2 ENSMUST00000024731.9 |
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr9_-_19163273 | 0.05 |
ENSMUST00000214019.2
ENSMUST00000214267.2 |
Olfr843
|
olfactory receptor 843 |
| chr6_+_37847721 | 0.05 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr4_+_62443606 | 0.05 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr2_-_89678487 | 0.05 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr10_+_40225272 | 0.05 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chrX_+_106299484 | 0.05 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr17_-_50600620 | 0.05 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr6_-_69704122 | 0.05 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr9_-_96601574 | 0.04 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr17_-_7620095 | 0.04 |
ENSMUST00000115747.3
|
Ttll2
|
tubulin tyrosine ligase-like family, member 2 |
| chrX_-_138683102 | 0.04 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr11_+_116734104 | 0.04 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr12_-_81579614 | 0.04 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr5_-_74692327 | 0.04 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr10_+_102210290 | 0.04 |
ENSMUST00000120748.2
|
Mgat4c
|
MGAT4 family, member C |
| chr11_-_94673526 | 0.04 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chr5_-_137213788 | 0.04 |
ENSMUST00000239135.2
|
Gm31160
|
predicted gene, 31160 |
| chr12_+_108572015 | 0.03 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr14_-_70666821 | 0.03 |
ENSMUST00000226229.2
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr18_+_34675366 | 0.03 |
ENSMUST00000012426.3
|
Wnt8a
|
wingless-type MMTV integration site family, member 8A |
| chr6_-_69394425 | 0.03 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr8_-_110688716 | 0.02 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr13_-_58506890 | 0.02 |
ENSMUST00000225388.2
|
Kif27
|
kinesin family member 27 |
| chr11_+_58668915 | 0.02 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr1_-_173161069 | 0.02 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr14_+_53521353 | 0.02 |
ENSMUST00000103625.3
|
Trav3n-3
|
T cell receptor alpha variable 3N-3 |
| chr7_-_44741609 | 0.02 |
ENSMUST00000210734.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr3_-_121056944 | 0.02 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr6_-_69245427 | 0.02 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chrX_-_9123138 | 0.02 |
ENSMUST00000115553.3
|
Gm14862
|
predicted gene 14862 |
| chr7_-_24371457 | 0.02 |
ENSMUST00000078001.7
|
Tex101
|
testis expressed gene 101 |
| chr9_-_105272435 | 0.02 |
ENSMUST00000140851.3
|
Nek11
|
NIMA (never in mitosis gene a)-related expressed kinase 11 |
| chr15_-_3333003 | 0.02 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr9_-_50472620 | 0.02 |
ENSMUST00000217236.2
|
Tex12
|
testis expressed 12 |
| chr9_+_40712562 | 0.01 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr18_+_12874368 | 0.01 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr14_-_52704952 | 0.01 |
ENSMUST00000206520.3
|
Olfr1508
|
olfactory receptor 1508 |
| chr9_-_50472605 | 0.01 |
ENSMUST00000034568.7
|
Tex12
|
testis expressed 12 |
| chr1_+_165288606 | 0.01 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr7_-_41098120 | 0.01 |
ENSMUST00000233793.2
ENSMUST00000233555.2 ENSMUST00000165029.3 |
Vmn2r57
|
vomeronasal 2, receptor 57 |
| chr18_+_12466876 | 0.01 |
ENSMUST00000092070.13
|
Lama3
|
laminin, alpha 3 |
| chr12_-_83534482 | 0.01 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr13_-_21726945 | 0.01 |
ENSMUST00000205976.3
ENSMUST00000175637.3 |
Olfr1366
|
olfactory receptor 1366 |
| chr14_+_25979825 | 0.01 |
ENSMUST00000173580.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr7_-_44741138 | 0.01 |
ENSMUST00000210527.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr16_-_20972750 | 0.01 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr7_+_106464189 | 0.01 |
ENSMUST00000098141.2
|
Olfr704
|
olfactory receptor 704 |
| chr7_-_139734637 | 0.01 |
ENSMUST00000059241.8
|
Sprn
|
shadow of prion protein |
| chr7_-_103420801 | 0.01 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr6_-_68994064 | 0.00 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr17_+_37716368 | 0.00 |
ENSMUST00000077008.4
|
Olfr107
|
olfactory receptor 107 |
| chr6_+_65019793 | 0.00 |
ENSMUST00000204696.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chrX_+_162694397 | 0.00 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr17_-_48145466 | 0.00 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr12_+_101370932 | 0.00 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr2_-_157188568 | 0.00 |
ENSMUST00000029172.2
|
Ghrh
|
growth hormone releasing hormone |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.3 | 0.8 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 1.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.6 | GO:0000239 | pachytene(GO:0000239) |
| 0.1 | 0.3 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.1 | 0.5 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.1 | 2.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.7 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.4 | GO:0014854 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.5 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 1.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.5 | GO:2000675 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 6.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 0.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 1.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |