Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxb4
Z-value: 0.28
Transcription factors associated with Hoxb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb4
|
ENSMUSG00000038692.9 | homeobox B4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb4 | mm39_v1_chr11_+_96209093_96209093 | 0.02 | 9.8e-01 | Click! |
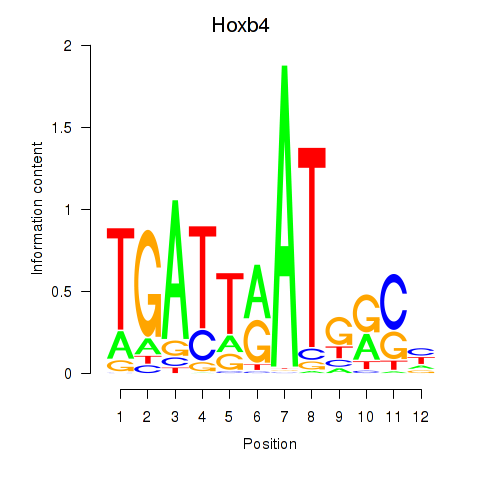
Activity profile of Hoxb4 motif
Sorted Z-values of Hoxb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_11063678 | 0.23 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr8_+_26298502 | 0.14 |
ENSMUST00000033979.6
|
Star
|
steroidogenic acute regulatory protein |
| chr2_-_84605732 | 0.12 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr18_-_56695333 | 0.08 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr2_-_84605764 | 0.07 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr1_-_171050004 | 0.06 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr18_+_69652880 | 0.06 |
ENSMUST00000200813.4
|
Tcf4
|
transcription factor 4 |
| chr6_+_142359099 | 0.06 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr7_-_119461027 | 0.06 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr7_+_28580847 | 0.05 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr8_-_106198112 | 0.05 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr18_+_69652837 | 0.04 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr10_-_89457115 | 0.04 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr2_-_118987305 | 0.04 |
ENSMUST00000135419.8
ENSMUST00000129351.2 ENSMUST00000139519.2 ENSMUST00000094695.12 |
Rmdn3
|
regulator of microtubule dynamics 3 |
| chrX_-_8059597 | 0.04 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr18_+_69652550 | 0.03 |
ENSMUST00000201205.4
|
Tcf4
|
transcription factor 4 |
| chr3_-_54621774 | 0.03 |
ENSMUST00000178832.2
|
Gm21958
|
predicted gene, 21958 |
| chrX_-_142716200 | 0.03 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr17_+_57586094 | 0.03 |
ENSMUST00000169220.9
ENSMUST00000005889.16 ENSMUST00000112870.5 |
Vav1
|
vav 1 oncogene |
| chr18_+_69652751 | 0.03 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chrX_+_165127688 | 0.03 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr4_+_43493344 | 0.02 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr8_-_23295603 | 0.02 |
ENSMUST00000163739.3
ENSMUST00000210656.2 |
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr18_+_56695515 | 0.02 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr10_+_96453408 | 0.02 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr3_-_20329823 | 0.02 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr9_-_60557076 | 0.02 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr3_-_51184730 | 0.02 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr17_+_44263890 | 0.01 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr13_+_94219934 | 0.01 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr11_-_40646090 | 0.01 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr12_-_54842488 | 0.01 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr1_-_171050077 | 0.01 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr7_-_29931612 | 0.01 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr18_-_56695288 | 0.00 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr2_+_102488985 | 0.00 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr18_-_56695259 | 0.00 |
ENSMUST00000171844.3
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr14_+_32972324 | 0.00 |
ENSMUST00000131086.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr16_-_94171340 | 0.00 |
ENSMUST00000138514.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_+_9615619 | 0.00 |
ENSMUST00000072079.9
|
Rrs1
|
ribosome biogenesis regulator 1 |
| chr9_+_39905108 | 0.00 |
ENSMUST00000057161.4
|
Olfr978
|
olfactory receptor 978 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.1 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |