Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxb6
Z-value: 0.36
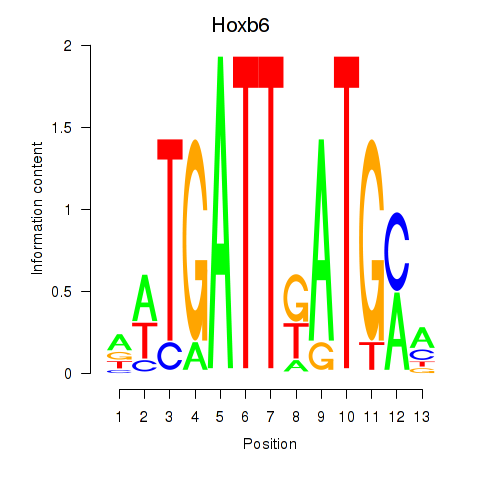
Transcription factors associated with Hoxb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb6
|
ENSMUSG00000000690.6 | homeobox B6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb6 | mm39_v1_chr11_+_96189963_96189997 | -0.91 | 3.2e-02 | Click! |
Activity profile of Hoxb6 motif
Sorted Z-values of Hoxb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_101943134 | 0.20 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr14_+_118374511 | 0.18 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr9_+_74959259 | 0.17 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr2_-_125701059 | 0.17 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr2_+_125701054 | 0.15 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chrX_+_37689503 | 0.15 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr3_-_100876960 | 0.14 |
ENSMUST00000076941.12
|
Ttf2
|
transcription termination factor, RNA polymerase II |
| chr1_-_138103021 | 0.13 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_72106418 | 0.13 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr8_+_111760521 | 0.13 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr8_-_94738748 | 0.12 |
ENSMUST00000143265.2
|
Amfr
|
autocrine motility factor receptor |
| chr1_-_138102972 | 0.12 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_74615496 | 0.11 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr10_-_117628565 | 0.11 |
ENSMUST00000167943.8
ENSMUST00000064848.7 |
Nup107
|
nucleoporin 107 |
| chr12_-_79211820 | 0.11 |
ENSMUST00000162569.8
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr2_+_85809620 | 0.09 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr3_+_68479578 | 0.09 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr6_-_119365632 | 0.08 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr4_-_134580604 | 0.07 |
ENSMUST00000030628.15
|
Maco1
|
macoilin 1 |
| chr9_-_56151334 | 0.07 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr5_+_88731366 | 0.07 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr9_-_35481689 | 0.07 |
ENSMUST00000115110.5
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr10_-_18103221 | 0.07 |
ENSMUST00000174592.8
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr3_-_51184730 | 0.06 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr13_-_118523760 | 0.05 |
ENSMUST00000022245.10
|
Mrps30
|
mitochondrial ribosomal protein S30 |
| chr8_-_78337297 | 0.05 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chrX_-_8118541 | 0.05 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chrX_+_106132055 | 0.04 |
ENSMUST00000150494.2
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr8_+_31601837 | 0.03 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr3_+_64884839 | 0.03 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_+_164611812 | 0.03 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr8_+_46452335 | 0.03 |
ENSMUST00000110380.8
ENSMUST00000066451.10 |
Lrp2bp
|
Lrp2 binding protein |
| chrX_+_56257374 | 0.03 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr16_+_44167484 | 0.02 |
ENSMUST00000050897.7
|
Spice1
|
spindle and centriole associated protein 1 |
| chr7_-_28661648 | 0.02 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr2_-_86589298 | 0.02 |
ENSMUST00000215991.2
ENSMUST00000217043.3 |
Olfr1090
|
olfactory receptor 1090 |
| chr15_+_34306812 | 0.02 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr2_-_86640362 | 0.01 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr10_-_38998272 | 0.01 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr12_-_119202527 | 0.01 |
ENSMUST00000026360.9
|
Itgb8
|
integrin beta 8 |
| chr6_-_3494587 | 0.01 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chrX_-_142716085 | 0.01 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chrX_+_162945162 | 0.01 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr19_+_12177998 | 0.01 |
ENSMUST00000213759.2
|
Olfr1431
|
olfactory receptor 1431 |
| chr1_+_106866678 | 0.01 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr19_-_6902698 | 0.01 |
ENSMUST00000237380.2
|
Catsperz
|
cation channel sperm associated auxiliary subunit zeta |
| chr3_+_94320548 | 0.01 |
ENSMUST00000166032.8
ENSMUST00000200486.5 ENSMUST00000196386.5 ENSMUST00000045245.10 ENSMUST00000197901.5 ENSMUST00000198041.2 |
Tdrkh
Gm42463
|
tudor and KH domain containing protein predicted gene 42463 |
| chr3_+_59989282 | 0.01 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr17_-_37481536 | 0.01 |
ENSMUST00000174673.3
|
Olfr753-ps1
|
olfactory receptor 753, pseudogene 1 |
| chr2_-_89193771 | 0.01 |
ENSMUST00000099783.2
|
Olfr1234
|
olfactory receptor 1234 |
| chr6_+_142244145 | 0.01 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr19_-_6902735 | 0.01 |
ENSMUST00000057716.6
|
Catsperz
|
cation channel sperm associated auxiliary subunit zeta |
| chr2_-_89328557 | 0.01 |
ENSMUST00000111540.4
ENSMUST00000216001.2 ENSMUST00000143935.3 |
Olfr1242
|
olfactory receptor 1242 |
| chr7_-_28661751 | 0.01 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr2_+_87284306 | 0.01 |
ENSMUST00000213366.3
|
Olfr1126
|
olfactory receptor 1126 |
| chr18_+_37864045 | 0.01 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr19_+_12245527 | 0.01 |
ENSMUST00000073507.3
|
Olfr235
|
olfactory receptor 235 |
| chr7_-_115459082 | 0.01 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_+_83642910 | 0.00 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr1_+_161222980 | 0.00 |
ENSMUST00000028024.5
|
Tnfsf4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr11_+_58471639 | 0.00 |
ENSMUST00000169428.5
|
Olfr325
|
olfactory receptor 325 |
| chr7_+_23702516 | 0.00 |
ENSMUST00000173571.2
|
V1rd19
|
vomeronasal 1 receptor, D19 |
| chr10_-_58511476 | 0.00 |
ENSMUST00000003312.5
|
Edar
|
ectodysplasin-A receptor |
| chr4_+_151081538 | 0.00 |
ENSMUST00000030803.2
|
Uts2
|
urotensin 2 |
| chr4_-_52936281 | 0.00 |
ENSMUST00000217546.2
|
Olfr271-ps1
|
olfactory receptor 271, pseudogene 1 |
| chr4_+_15881256 | 0.00 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chr2_-_89983396 | 0.00 |
ENSMUST00000215578.2
ENSMUST00000216354.2 ENSMUST00000217139.2 ENSMUST00000213946.3 ENSMUST00000215975.3 |
Olfr1270
|
olfactory receptor 1270 |
| chr7_+_108019628 | 0.00 |
ENSMUST00000213521.3
|
Olfr497
|
olfactory receptor 497 |
| chr7_+_112806672 | 0.00 |
ENSMUST00000047321.9
ENSMUST00000210074.2 ENSMUST00000210238.2 |
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr1_-_36312482 | 0.00 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr11_-_58393380 | 0.00 |
ENSMUST00000170501.3
ENSMUST00000081743.3 |
Olfr331
|
olfactory receptor 331 |
| chr19_+_12405268 | 0.00 |
ENSMUST00000168148.2
|
Pfpl
|
pore forming protein-like |
| chr14_-_52728503 | 0.00 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
| chr6_-_69377328 | 0.00 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr11_-_69686756 | 0.00 |
ENSMUST00000045971.9
|
Chrnb1
|
cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) |
| chr1_+_173097714 | 0.00 |
ENSMUST00000059754.3
|
Olfr418
|
olfactory receptor 418 |
| chr1_-_56676589 | 0.00 |
ENSMUST00000062085.6
|
Hsfy2
|
heat shock transcription factor, Y-linked 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |