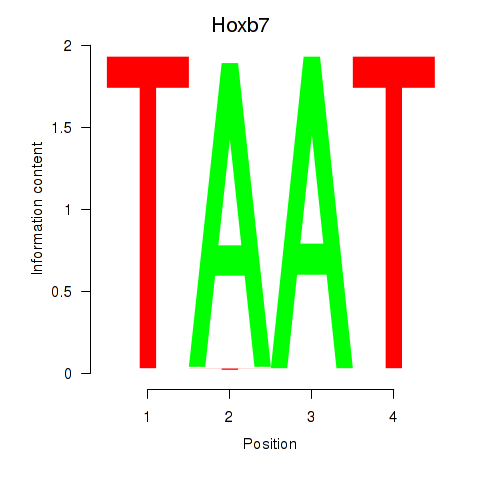
| 0.2 |
1.2 |
GO:0003409 |
optic cup structural organization(GO:0003409) |
| 0.2 |
0.9 |
GO:1904799 |
regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.2 |
1.3 |
GO:1902748 |
positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 |
2.1 |
GO:0003185 |
primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 |
0.7 |
GO:0002358 |
B cell homeostatic proliferation(GO:0002358) |
| 0.1 |
0.3 |
GO:2000729 |
positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 |
0.5 |
GO:1905051 |
regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 |
0.6 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.1 |
0.5 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.5 |
GO:0021502 |
neural fold elevation formation(GO:0021502) |
| 0.1 |
0.8 |
GO:0007527 |
adult somatic muscle development(GO:0007527) |
| 0.1 |
0.6 |
GO:0071698 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.4 |
GO:0060133 |
somatotropin secreting cell development(GO:0060133) |
| 0.1 |
0.3 |
GO:1900108 |
sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 |
0.3 |
GO:0045358 |
negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 |
0.4 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 |
0.3 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.1 |
0.1 |
GO:0050787 |
detoxification of mercury ion(GO:0050787) |
| 0.1 |
0.3 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.1 |
0.2 |
GO:0097374 |
sensory neuron axon guidance(GO:0097374) |
| 0.1 |
0.2 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
0.2 |
GO:1904956 |
regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 |
0.4 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.2 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.3 |
GO:0097168 |
mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 |
0.2 |
GO:0034378 |
chylomicron assembly(GO:0034378) |
| 0.1 |
0.2 |
GO:0043380 |
regulation of memory T cell differentiation(GO:0043380) |
| 0.1 |
0.2 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.1 |
0.3 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.1 |
0.3 |
GO:0021553 |
olfactory nerve development(GO:0021553) |
| 0.1 |
0.5 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 |
0.4 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.1 |
0.2 |
GO:0050973 |
detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 |
0.3 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 |
0.2 |
GO:0070904 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.1 |
GO:0001743 |
optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 |
0.8 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 |
0.2 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.2 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 |
0.2 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.3 |
GO:0002023 |
reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 |
0.1 |
GO:0051795 |
positive regulation of catagen(GO:0051795) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 |
0.5 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 0.0 |
1.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.1 |
GO:0002305 |
gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 |
0.2 |
GO:0055048 |
regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 |
0.2 |
GO:0036060 |
filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
1.0 |
GO:0060044 |
negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 |
0.2 |
GO:0023016 |
signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 |
0.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.0 |
GO:0060595 |
fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 |
0.2 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.0 |
0.3 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.3 |
GO:0086023 |
adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 |
0.3 |
GO:0035865 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 |
0.2 |
GO:1901295 |
Notch signaling pathway involved in heart induction(GO:0003137) regulation of Notch signaling pathway involved in heart induction(GO:0035480) positive regulation of Notch signaling pathway involved in heart induction(GO:0035481) regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 |
0.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.1 |
GO:0070563 |
cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 |
0.2 |
GO:1904970 |
brush border assembly(GO:1904970) |
| 0.0 |
0.2 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.0 |
0.2 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.1 |
GO:0034635 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 |
0.3 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.5 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 |
0.1 |
GO:0044337 |
canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 |
0.1 |
GO:0061146 |
Peyer's patch morphogenesis(GO:0061146) |
| 0.0 |
0.3 |
GO:0036309 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.6 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.2 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.1 |
GO:0060061 |
Spemann organizer formation(GO:0060061) |
| 0.0 |
0.1 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.1 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.0 |
0.1 |
GO:0098884 |
postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 |
0.3 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.1 |
GO:0007208 |
phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 |
0.1 |
GO:0009305 |
protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 |
0.3 |
GO:0045964 |
positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 |
0.1 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.0 |
0.3 |
GO:0038044 |
transforming growth factor-beta secretion(GO:0038044) |
| 0.0 |
0.2 |
GO:0035127 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 |
0.1 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.0 |
0.2 |
GO:1903566 |
positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 |
0.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.5 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.1 |
GO:0021633 |
optic nerve structural organization(GO:0021633) |
| 0.0 |
0.1 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
0.5 |
GO:0002664 |
regulation of T cell tolerance induction(GO:0002664) |
| 0.0 |
0.0 |
GO:0072069 |
DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) |
| 0.0 |
0.1 |
GO:0055005 |
ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 |
0.3 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.2 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 |
0.1 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.0 |
0.3 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.1 |
GO:1904428 |
negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 |
0.1 |
GO:1904075 |
regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 |
0.1 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 |
0.2 |
GO:0021842 |
directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 |
0.0 |
GO:0014737 |
positive regulation of muscle atrophy(GO:0014737) |
| 0.0 |
0.2 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.0 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 |
0.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.1 |
GO:0051935 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 |
0.1 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.1 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.0 |
0.1 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.1 |
GO:0070173 |
regulation of enamel mineralization(GO:0070173) |
| 0.0 |
0.1 |
GO:0097476 |
spinal cord motor neuron migration(GO:0097476) |
| 0.0 |
0.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.1 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.0 |
0.1 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.1 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.1 |
GO:0021776 |
smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 |
0.1 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 |
0.0 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0002729 |
positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 |
0.2 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.0 |
GO:1902164 |
positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 |
0.1 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:0070495 |
regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 |
0.0 |
GO:0002414 |
immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 |
0.6 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 |
0.0 |
GO:0046874 |
quinolinate metabolic process(GO:0046874) |
| 0.0 |
0.1 |
GO:0060139 |
positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
0.1 |
GO:0098957 |
anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 |
0.4 |
GO:0042438 |
melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.0 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 |
0.1 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 |
0.1 |
GO:0086018 |
SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.0 |
0.1 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.0 |
0.1 |
GO:2000359 |
regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 |
0.2 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.1 |
GO:0035585 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 |
0.1 |
GO:1903919 |
regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.0 |
0.1 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.0 |
0.1 |
GO:0046604 |
positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 |
0.3 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.6 |
GO:0050911 |
detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.0 |
0.0 |
GO:0061536 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0075509 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.1 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.3 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.1 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.0 |
0.1 |
GO:1904016 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 |
0.2 |
GO:0044154 |
histone H3-K14 acetylation(GO:0044154) |
| 0.0 |
0.2 |
GO:0097152 |
mesenchymal cell apoptotic process(GO:0097152) regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.0 |
GO:0038016 |
insulin receptor internalization(GO:0038016) |
| 0.0 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.0 |
GO:1905154 |
negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 |
0.1 |
GO:0033183 |
negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 |
0.0 |
GO:0000101 |
sulfur amino acid transport(GO:0000101) |
| 0.0 |
0.1 |
GO:0071313 |
cellular response to caffeine(GO:0071313) |
| 0.0 |
0.1 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 |
0.1 |
GO:0002826 |
negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 |
0.0 |
GO:0045226 |
extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 |
0.4 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.0 |
0.1 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.1 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:0010255 |
carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 |
0.4 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.0 |
GO:0002632 |
regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0009996 |
negative regulation of cell fate specification(GO:0009996) |
| 0.0 |
0.1 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.2 |
GO:0098953 |
receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 |
0.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.2 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.1 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.0 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.0 |
0.1 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.0 |
0.4 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.1 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 |
0.1 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 |
0.0 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |