Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxb8_Pdx1
Z-value: 0.38
Transcription factors associated with Hoxb8_Pdx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
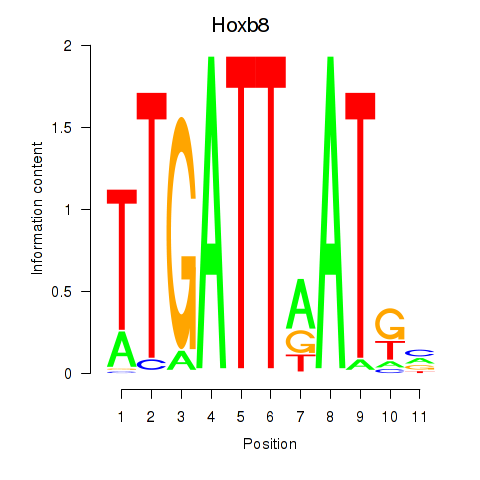
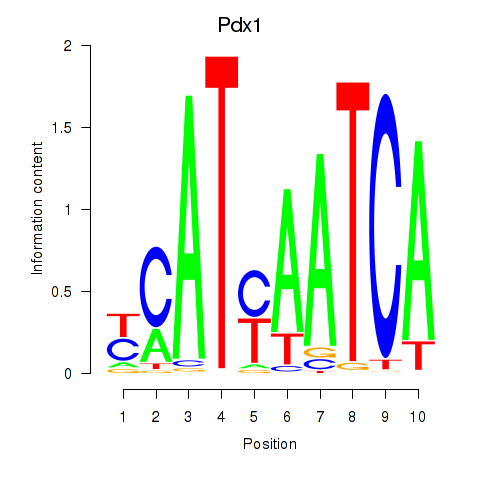
Hoxb8
|
ENSMUSG00000056648.6 | homeobox B8 |
|
Pdx1
|
ENSMUSG00000029644.8 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pdx1 | mm39_v1_chr5_+_147206769_147206912 | -0.25 | 6.8e-01 | Click! |
Activity profile of Hoxb8_Pdx1 motif
Sorted Z-values of Hoxb8_Pdx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_4829519 | 0.37 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr6_-_52141796 | 0.35 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr14_-_31503869 | 0.34 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr8_-_111629074 | 0.31 |
ENSMUST00000041382.7
|
Fcsk
|
fucose kinase |
| chr8_-_4829473 | 0.27 |
ENSMUST00000207262.2
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr13_+_94954202 | 0.24 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr3_-_65299967 | 0.22 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr3_-_65300000 | 0.22 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr11_+_49138278 | 0.20 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chrX_+_37689503 | 0.19 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr10_+_96453408 | 0.17 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr13_+_24986003 | 0.17 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr1_-_120197979 | 0.17 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr7_-_27749453 | 0.15 |
ENSMUST00000140053.3
ENSMUST00000032824.10 |
Psmc4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr14_+_74973081 | 0.15 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr19_-_24178000 | 0.15 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr3_+_121243395 | 0.14 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr7_-_99512558 | 0.13 |
ENSMUST00000207137.2
ENSMUST00000207063.2 ENSMUST00000207580.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr1_-_120198804 | 0.12 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr8_+_12999480 | 0.12 |
ENSMUST00000110866.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr1_-_149798090 | 0.12 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr12_-_75678092 | 0.12 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr19_-_32443978 | 0.12 |
ENSMUST00000078034.5
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr10_+_42554888 | 0.12 |
ENSMUST00000040718.6
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr2_+_36120438 | 0.12 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr16_+_4756963 | 0.12 |
ENSMUST00000229961.2
ENSMUST00000050881.10 |
Nudt16l1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 |
| chr2_+_174127049 | 0.11 |
ENSMUST00000130940.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr18_-_35631914 | 0.11 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr4_-_150087587 | 0.10 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr6_-_30390996 | 0.10 |
ENSMUST00000152391.9
ENSMUST00000115184.2 ENSMUST00000080812.14 ENSMUST00000102992.10 |
Zc3hc1
|
zinc finger, C3HC type 1 |
| chr2_-_151586063 | 0.10 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr19_+_24853039 | 0.10 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr4_-_41723129 | 0.10 |
ENSMUST00000171641.2
ENSMUST00000030158.11 |
Dctn3
|
dynactin 3 |
| chr5_+_30360246 | 0.10 |
ENSMUST00000026841.15
ENSMUST00000123980.8 ENSMUST00000114783.6 ENSMUST00000114786.8 |
Hadhb
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr8_+_12999394 | 0.09 |
ENSMUST00000110867.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr9_+_121539395 | 0.09 |
ENSMUST00000035113.11
ENSMUST00000215966.2 ENSMUST00000215833.2 ENSMUST00000215104.2 |
Ss18l2
|
SS18, nBAF chromatin remodeling complex subunit like 2 |
| chrX_-_36137764 | 0.09 |
ENSMUST00000047486.6
|
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr7_-_125968653 | 0.09 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr12_-_79211820 | 0.09 |
ENSMUST00000162569.8
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr6_-_52185674 | 0.09 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr3_-_37778470 | 0.09 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr2_-_155668567 | 0.09 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr9_-_31043076 | 0.09 |
ENSMUST00000034478.3
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr2_-_125701059 | 0.08 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr1_-_37575313 | 0.08 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr7_+_130633776 | 0.08 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr4_+_146586445 | 0.08 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr18_+_77017713 | 0.08 |
ENSMUST00000026487.6
ENSMUST00000136800.2 |
Ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr6_-_119365632 | 0.08 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr1_+_134487928 | 0.08 |
ENSMUST00000112198.3
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr15_-_77037972 | 0.07 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_+_87490666 | 0.07 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr14_+_26722319 | 0.07 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr7_+_46490899 | 0.07 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr13_+_93440572 | 0.07 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chrM_+_8603 | 0.07 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr16_-_65359566 | 0.07 |
ENSMUST00000004965.8
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr11_+_94218810 | 0.07 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr3_-_51184895 | 0.07 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr6_+_88061464 | 0.07 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr18_-_66155651 | 0.07 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr13_+_93441307 | 0.07 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr16_+_49620883 | 0.07 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr14_+_55797468 | 0.07 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chrX_-_8118541 | 0.07 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr5_-_109706801 | 0.07 |
ENSMUST00000200284.5
ENSMUST00000044579.12 |
Crlf2
|
cytokine receptor-like factor 2 |
| chrM_+_9459 | 0.07 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr12_-_25147139 | 0.07 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr4_+_107747010 | 0.07 |
ENSMUST00000135454.8
ENSMUST00000106726.10 ENSMUST00000106727.10 ENSMUST00000119394.8 ENSMUST00000120473.8 ENSMUST00000125107.8 ENSMUST00000128474.2 |
Czib
|
CXXC motif containing zinc binding protein |
| chr1_+_88062508 | 0.07 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr16_-_65359406 | 0.07 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr3_-_51184730 | 0.07 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr9_+_98873831 | 0.07 |
ENSMUST00000185472.2
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr14_+_55797443 | 0.06 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr2_+_125701054 | 0.06 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr9_-_39405284 | 0.06 |
ENSMUST00000077757.7
ENSMUST00000215065.3 ENSMUST00000216316.3 |
Olfr44
|
olfactory receptor 44 |
| chr14_+_48683797 | 0.06 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr6_-_52203146 | 0.06 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr13_-_96217879 | 0.06 |
ENSMUST00000220449.2
|
Gm20075
|
predicted gene, 20075 |
| chr18_+_3383230 | 0.06 |
ENSMUST00000162301.8
ENSMUST00000161317.2 |
Cul2
|
cullin 2 |
| chr1_+_87983099 | 0.06 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr4_-_115932219 | 0.06 |
ENSMUST00000050580.11
ENSMUST00000078676.6 |
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr15_+_31224555 | 0.06 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr15_+_102381929 | 0.06 |
ENSMUST00000229746.2
ENSMUST00000230211.2 ENSMUST00000230539.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr18_-_24153363 | 0.06 |
ENSMUST00000153337.2
ENSMUST00000148525.2 |
Zfp24
|
zinc finger protein 24 |
| chr13_-_21685588 | 0.06 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr18_-_36916148 | 0.06 |
ENSMUST00000001416.8
|
Hars
|
histidyl-tRNA synthetase |
| chr3_+_151143524 | 0.06 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr7_+_45683122 | 0.06 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr16_-_56533179 | 0.05 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr1_-_189654535 | 0.05 |
ENSMUST00000027897.8
|
Smyd2
|
SET and MYND domain containing 2 |
| chr17_-_35265702 | 0.05 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr14_+_55798362 | 0.05 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_-_130053120 | 0.05 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr11_-_74615496 | 0.05 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr15_-_42540363 | 0.05 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr5_-_30360113 | 0.05 |
ENSMUST00000156859.3
|
Hadha
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| chr11_-_121095462 | 0.05 |
ENSMUST00000026169.7
|
Ogfod3
|
2-oxoglutarate and iron-dependent oxygenase domain containing 3 |
| chr11_+_103024128 | 0.05 |
ENSMUST00000107037.8
ENSMUST00000124928.2 ENSMUST00000062530.5 |
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr17_-_33879224 | 0.05 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr4_+_127137577 | 0.05 |
ENSMUST00000142029.2
|
Smim12
|
small integral membrane protein 12 |
| chr12_+_103354915 | 0.05 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr16_+_35803794 | 0.05 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr17_-_56447332 | 0.05 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr15_+_21111428 | 0.05 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr12_-_65010124 | 0.05 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr17_+_24645615 | 0.05 |
ENSMUST00000234304.2
ENSMUST00000024946.7 ENSMUST00000234150.2 |
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr17_+_6130205 | 0.05 |
ENSMUST00000100955.3
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr17_-_71309815 | 0.05 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr15_+_102381705 | 0.05 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr1_-_125362321 | 0.05 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr17_-_35081129 | 0.05 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr6_-_129252396 | 0.05 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr1_-_164763091 | 0.05 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr11_-_4897991 | 0.04 |
ENSMUST00000093369.5
|
Nefh
|
neurofilament, heavy polypeptide |
| chr7_-_99994257 | 0.04 |
ENSMUST00000207634.2
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr6_+_106095726 | 0.04 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr17_+_6130061 | 0.04 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr8_-_117809188 | 0.04 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr13_+_19398273 | 0.04 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr16_+_20430335 | 0.04 |
ENSMUST00000115522.10
ENSMUST00000119224.8 ENSMUST00000120394.8 ENSMUST00000079600.12 |
Eef1akmt4
Gm49333
|
EEF1A lysine methyltransferase 4 predicted gene, 49333 |
| chr2_-_69036489 | 0.04 |
ENSMUST00000127243.8
ENSMUST00000149643.2 ENSMUST00000167875.9 ENSMUST00000005365.15 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr3_+_89173859 | 0.04 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr2_-_69036472 | 0.04 |
ENSMUST00000112320.8
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr11_-_23583591 | 0.04 |
ENSMUST00000180260.8
ENSMUST00000141353.8 ENSMUST00000131612.2 ENSMUST00000109532.9 |
0610010F05Rik
|
RIKEN cDNA 0610010F05 gene |
| chr15_+_31225302 | 0.04 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr15_-_36496880 | 0.04 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr1_+_87983189 | 0.04 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_106890307 | 0.04 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr1_-_63215812 | 0.04 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr9_+_35470752 | 0.04 |
ENSMUST00000034615.10
ENSMUST00000121246.2 |
Pus3
|
pseudouridine synthase 3 |
| chr1_-_185061525 | 0.04 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr2_-_38895586 | 0.04 |
ENSMUST00000080861.6
|
Rpl35
|
ribosomal protein L35 |
| chr12_-_54842488 | 0.04 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr17_-_35081456 | 0.04 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr5_+_38233901 | 0.04 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr18_+_47245204 | 0.04 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr2_-_84255602 | 0.04 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr18_+_56695515 | 0.04 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr6_+_57679455 | 0.04 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr11_-_72106418 | 0.04 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr10_-_18103221 | 0.04 |
ENSMUST00000174592.8
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr4_+_109092459 | 0.03 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr8_-_68270870 | 0.03 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_-_129107354 | 0.03 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr6_+_115578863 | 0.03 |
ENSMUST00000000449.9
|
Mkrn2
|
makorin, ring finger protein, 2 |
| chr15_+_52575796 | 0.03 |
ENSMUST00000037115.9
|
Med30
|
mediator complex subunit 30 |
| chr3_+_89337568 | 0.03 |
ENSMUST00000060061.11
|
Pygo2
|
pygopus 2 |
| chr12_+_117621644 | 0.03 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr8_-_84059048 | 0.03 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr17_+_29170924 | 0.03 |
ENSMUST00000153462.8
|
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr6_+_57679621 | 0.03 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr10_+_4561974 | 0.03 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr7_-_126641565 | 0.03 |
ENSMUST00000205806.2
|
Kif22
|
kinesin family member 22 |
| chr1_+_134487893 | 0.03 |
ENSMUST00000047714.14
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr14_+_55797934 | 0.03 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_+_130285870 | 0.03 |
ENSMUST00000100662.10
ENSMUST00000040213.13 |
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr18_-_32044877 | 0.03 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr13_+_94219934 | 0.03 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr8_-_106292616 | 0.03 |
ENSMUST00000013304.8
|
Atp6v0d1
|
ATPase, H+ transporting, lysosomal V0 subunit D1 |
| chrX_+_20529137 | 0.03 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr10_-_14581203 | 0.03 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr2_+_152873772 | 0.03 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr2_-_151583155 | 0.03 |
ENSMUST00000042452.11
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr11_-_103829040 | 0.03 |
ENSMUST00000133774.4
ENSMUST00000149642.3 |
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr3_-_129548954 | 0.03 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr9_-_13245834 | 0.03 |
ENSMUST00000110582.4
|
Jrkl
|
Jrk-like |
| chr15_+_5215000 | 0.03 |
ENSMUST00000118193.8
ENSMUST00000022751.15 |
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr5_+_117378510 | 0.03 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr4_+_148985584 | 0.03 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chr3_+_32583602 | 0.03 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr1_-_144651157 | 0.03 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr15_-_77037926 | 0.03 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr4_+_147576874 | 0.03 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr17_+_25517363 | 0.03 |
ENSMUST00000037453.4
|
Prss34
|
protease, serine 34 |
| chr9_+_35334878 | 0.03 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr18_+_38430205 | 0.03 |
ENSMUST00000235491.2
|
Rnf14
|
ring finger protein 14 |
| chr18_+_37898633 | 0.03 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr4_+_108336164 | 0.03 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr3_-_103553347 | 0.03 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr10_-_116385007 | 0.03 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr11_+_78403019 | 0.03 |
ENSMUST00000001127.11
|
Poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr7_-_4815111 | 0.03 |
ENSMUST00000205885.2
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr8_+_85065268 | 0.03 |
ENSMUST00000238701.2
|
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr8_-_68270936 | 0.03 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_-_4793274 | 0.03 |
ENSMUST00000084949.3
|
Bpnt2
|
3'(2'), 5'-bisphosphate nucleotidase 2 |
| chr12_-_114012399 | 0.03 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr6_+_21949570 | 0.03 |
ENSMUST00000031680.10
ENSMUST00000115389.8 ENSMUST00000151473.8 |
Ing3
|
inhibitor of growth family, member 3 |
| chr9_-_71070506 | 0.03 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr5_-_38649291 | 0.03 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr1_+_88030951 | 0.03 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr3_+_68776884 | 0.03 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr15_+_82230155 | 0.03 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr18_+_36916272 | 0.03 |
ENSMUST00000019287.9
|
Hars2
|
histidyl-tRNA synthetase 2 |
| chr4_+_145397238 | 0.03 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr1_-_45542442 | 0.03 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr7_-_144837719 | 0.03 |
ENSMUST00000058022.6
|
Tpcn2
|
two pore segment channel 2 |
| chrX_-_142716085 | 0.02 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr10_+_5543769 | 0.02 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr8_-_106223502 | 0.02 |
ENSMUST00000212303.2
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb8_Pdx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.3 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:2000556 | immunoglobulin production in mucosal tissue(GO:0002426) positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |