Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxc10
Z-value: 1.14
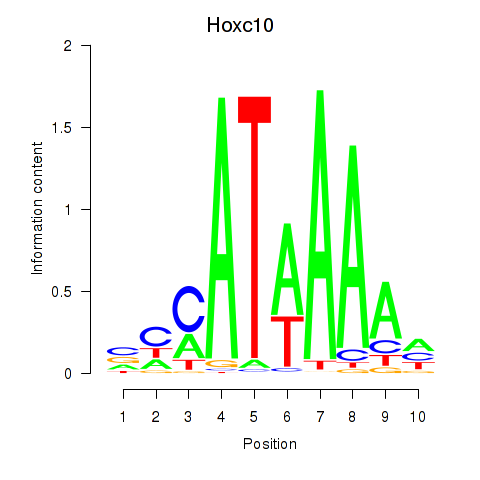
Transcription factors associated with Hoxc10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc10
|
ENSMUSG00000022484.8 | homeobox C10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc10 | mm39_v1_chr15_+_102875229_102875248 | -0.74 | 1.5e-01 | Click! |
Activity profile of Hoxc10 motif
Sorted Z-values of Hoxc10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_151586063 | 1.32 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr14_-_31503869 | 1.06 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr11_+_96085118 | 0.89 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chrX_-_133442596 | 0.88 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr2_+_80145805 | 0.87 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr10_-_35587888 | 0.85 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr13_+_94954202 | 0.84 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chrX_+_138464065 | 0.79 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr7_-_30428930 | 0.76 |
ENSMUST00000207296.2
ENSMUST00000006478.10 |
Tmem147
|
transmembrane protein 147 |
| chr11_+_52265090 | 0.72 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chrX_+_55493325 | 0.68 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr4_-_154721288 | 0.66 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr1_-_149836974 | 0.65 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr1_-_45542442 | 0.58 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr7_-_30428746 | 0.55 |
ENSMUST00000209065.2
ENSMUST00000208169.2 |
Tmem147
|
transmembrane protein 147 |
| chr15_-_78947038 | 0.54 |
ENSMUST00000151889.8
ENSMUST00000040676.11 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr2_-_32278245 | 0.51 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr14_+_66043281 | 0.47 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr3_-_146475974 | 0.44 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr9_+_113641615 | 0.43 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr11_-_120440519 | 0.43 |
ENSMUST00000034913.5
|
Mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr2_-_86180622 | 0.41 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr2_-_32341408 | 0.40 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chrX_+_168468186 | 0.39 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr2_-_69542805 | 0.38 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr1_-_97689263 | 0.38 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr13_+_38223023 | 0.36 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr15_+_25774070 | 0.34 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr1_-_144124871 | 0.33 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr11_-_61268879 | 0.32 |
ENSMUST00000010267.10
|
Slc47a1
|
solute carrier family 47, member 1 |
| chr1_+_179788037 | 0.29 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr11_-_102210568 | 0.29 |
ENSMUST00000173870.8
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr4_+_11758147 | 0.27 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr3_-_88317601 | 0.27 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr6_-_52203146 | 0.25 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr12_+_117621644 | 0.23 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr11_+_94102255 | 0.22 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr2_+_73102269 | 0.22 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr8_-_94739469 | 0.22 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr19_+_56536685 | 0.21 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr10_+_87926932 | 0.20 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr1_+_179788675 | 0.18 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr13_+_104424359 | 0.18 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr1_-_171854818 | 0.18 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr4_+_118217179 | 0.18 |
ENSMUST00000006562.6
|
Hyi
|
hydroxypyruvate isomerase (putative) |
| chr15_+_6451721 | 0.18 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr8_-_47805383 | 0.17 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr19_-_56536443 | 0.17 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr9_+_37119472 | 0.16 |
ENSMUST00000034632.10
|
Tmem218
|
transmembrane protein 218 |
| chr11_+_108811168 | 0.15 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr11_+_96214078 | 0.15 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr1_+_61017057 | 0.14 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr3_+_89173859 | 0.14 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr8_-_41469332 | 0.14 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_129213050 | 0.13 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr1_-_36283326 | 0.13 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr13_+_25127127 | 0.13 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr19_-_56536646 | 0.12 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr15_+_21111428 | 0.12 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr7_-_141014477 | 0.12 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr10_-_6930376 | 0.12 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr17_+_34043536 | 0.11 |
ENSMUST00000048249.8
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr9_+_53678801 | 0.11 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr11_-_73215442 | 0.11 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr11_+_100902572 | 0.09 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr8_-_62576140 | 0.09 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr14_+_54923655 | 0.08 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr16_+_41353360 | 0.08 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr5_+_143864993 | 0.07 |
ENSMUST00000172367.3
|
Gm42421
|
predicted gene, 42421 |
| chr1_+_189460461 | 0.07 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr19_-_50667079 | 0.07 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr1_-_144124787 | 0.07 |
ENSMUST00000185714.2
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr10_-_89270554 | 0.06 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr3_-_63758672 | 0.06 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chrX_+_95498965 | 0.06 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr18_-_3309858 | 0.06 |
ENSMUST00000144496.8
ENSMUST00000154715.8 |
Crem
|
cAMP responsive element modulator |
| chr10_-_28862289 | 0.06 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr15_+_91722524 | 0.06 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr18_+_44249254 | 0.05 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797 |
| chr15_-_101389384 | 0.05 |
ENSMUST00000023718.9
|
Krt83
|
keratin 83 |
| chr11_+_108811626 | 0.05 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr5_-_86437119 | 0.05 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr13_-_21765822 | 0.05 |
ENSMUST00000206526.3
ENSMUST00000213912.2 |
Olfr1364
|
olfactory receptor 1364 |
| chr10_+_52267702 | 0.05 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr12_-_113912416 | 0.05 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr5_+_90708962 | 0.05 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr3_-_49711706 | 0.04 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr15_-_101422054 | 0.04 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr10_-_89270527 | 0.04 |
ENSMUST00000218764.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr2_+_177783713 | 0.04 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr3_-_49711765 | 0.04 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr11_-_99877423 | 0.04 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr17_+_31652029 | 0.04 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr18_-_3309723 | 0.04 |
ENSMUST00000152108.8
ENSMUST00000136961.8 |
Crem
|
cAMP responsive element modulator |
| chr1_+_66426127 | 0.04 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr18_+_20443795 | 0.04 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr17_-_34043502 | 0.04 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr14_+_27598021 | 0.04 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr4_-_96673423 | 0.04 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr11_-_115824290 | 0.04 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr17_-_38442081 | 0.04 |
ENSMUST00000087128.2
|
Olfr132
|
olfactory receptor 132 |
| chr15_-_101441254 | 0.04 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr17_+_31652073 | 0.03 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr7_+_107501834 | 0.03 |
ENSMUST00000210420.2
|
Olfr472
|
olfactory receptor 472 |
| chr10_-_38998272 | 0.03 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr16_-_88759753 | 0.03 |
ENSMUST00000179707.3
|
Krtap16-3
|
keratin associated protein 16-3 |
| chr7_-_30312246 | 0.03 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A |
| chr9_-_44473146 | 0.03 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr9_-_15023396 | 0.03 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr3_-_122828592 | 0.03 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr17_-_91400142 | 0.03 |
ENSMUST00000160800.9
|
Nrxn1
|
neurexin I |
| chr5_+_88523967 | 0.03 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr8_+_94537910 | 0.03 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr11_-_99996452 | 0.03 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr6_-_136852792 | 0.03 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr1_+_107439145 | 0.03 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr15_+_91722458 | 0.03 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr6_-_133014064 | 0.03 |
ENSMUST00000032317.4
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr4_+_102111936 | 0.03 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr17_-_35293447 | 0.03 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr16_-_89305201 | 0.03 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr2_-_86938729 | 0.03 |
ENSMUST00000099862.2
|
Olfr259
|
olfactory receptor 259 |
| chr2_+_83642910 | 0.02 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr10_-_116732813 | 0.02 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr10_-_129023288 | 0.02 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chr19_+_39102342 | 0.02 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr3_+_31956656 | 0.02 |
ENSMUST00000119310.8
|
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr16_+_26281885 | 0.02 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr16_-_19385631 | 0.02 |
ENSMUST00000090062.2
|
Olfr169
|
olfactory receptor 169 |
| chr5_+_149942140 | 0.02 |
ENSMUST00000065745.10
ENSMUST00000110496.5 ENSMUST00000201612.2 |
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr6_-_24168082 | 0.02 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr10_+_90412827 | 0.02 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_-_99884818 | 0.02 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr11_-_99932373 | 0.02 |
ENSMUST00000056362.3
|
Krt34
|
keratin 34 |
| chr2_-_87805007 | 0.02 |
ENSMUST00000099840.2
|
Olfr74
|
olfactory receptor 74 |
| chr16_-_19443851 | 0.02 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr2_+_65499097 | 0.02 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr7_-_115459082 | 0.02 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr11_-_83957889 | 0.02 |
ENSMUST00000108101.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr2_-_86521317 | 0.02 |
ENSMUST00000099877.2
|
Olfr1087
|
olfactory receptor 1087 |
| chr16_-_19425453 | 0.02 |
ENSMUST00000078603.3
ENSMUST00000218837.2 |
Olfr170
|
olfactory receptor 170 |
| chr11_-_55498559 | 0.02 |
ENSMUST00000108853.8
ENSMUST00000075603.5 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr2_-_35994819 | 0.02 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr15_+_92495007 | 0.02 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr2_+_142920386 | 0.02 |
ENSMUST00000028902.3
|
Otor
|
otoraplin |
| chr10_+_69816153 | 0.02 |
ENSMUST00000182207.8
|
Ank3
|
ankyrin 3, epithelial |
| chr15_-_101459092 | 0.01 |
ENSMUST00000023713.10
|
Krt82
|
keratin 82 |
| chr15_-_101482320 | 0.01 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr13_+_83723255 | 0.01 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr18_+_20380397 | 0.01 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr2_+_163348728 | 0.01 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr7_-_104856692 | 0.01 |
ENSMUST00000216143.2
|
Olfr686
|
olfactory receptor 686 |
| chr2_-_89980380 | 0.01 |
ENSMUST00000099754.5
|
Olfr1270
|
olfactory receptor 1270 |
| chr3_+_31956814 | 0.01 |
ENSMUST00000192429.6
ENSMUST00000191869.6 ENSMUST00000178668.2 ENSMUST00000119970.8 |
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr10_+_23875837 | 0.01 |
ENSMUST00000092658.2
|
Taar7b
|
trace amine-associated receptor 7B |
| chr5_-_67256578 | 0.01 |
ENSMUST00000012664.11
|
Phox2b
|
paired-like homeobox 2b |
| chr11_-_100036792 | 0.01 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr16_-_58749007 | 0.01 |
ENSMUST00000075361.5
ENSMUST00000205668.2 ENSMUST00000205986.4 |
Olfr181
|
olfactory receptor 181 |
| chr7_-_30623592 | 0.01 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr7_-_29605504 | 0.01 |
ENSMUST00000053521.15
|
Zfp27
|
zinc finger protein 27 |
| chr4_+_128653051 | 0.01 |
ENSMUST00000030585.8
|
A3galt2
|
alpha 1,3-galactosyltransferase 2 (isoglobotriaosylceramide synthase) |
| chr3_-_14843512 | 0.01 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chrX_-_166907286 | 0.01 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr7_-_103261741 | 0.01 |
ENSMUST00000052152.3
|
Olfr620
|
olfactory receptor 620 |
| chr7_-_102604827 | 0.01 |
ENSMUST00000084812.3
|
Olfr575
|
olfactory receptor 575 |
| chrX_+_132809189 | 0.01 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr12_+_105570350 | 0.01 |
ENSMUST00000041229.5
|
Bdkrb1
|
bradykinin receptor, beta 1 |
| chr1_-_69147185 | 0.01 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr5_+_43673093 | 0.00 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr16_+_29884153 | 0.00 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr10_-_49659355 | 0.00 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr10_+_23796254 | 0.00 |
ENSMUST00000051532.5
|
Taar1
|
trace amine-associated receptor 1 |
| chr2_-_121381832 | 0.00 |
ENSMUST00000212518.2
|
Frmd5
|
FERM domain containing 5 |
| chr13_-_18347125 | 0.00 |
ENSMUST00000184299.2
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr10_-_33920289 | 0.00 |
ENSMUST00000218239.2
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr16_-_38620688 | 0.00 |
ENSMUST00000057767.6
|
Upk1b
|
uroplakin 1B |
| chr8_-_96301825 | 0.00 |
ENSMUST00000180075.2
|
Prss54
|
protease, serine 54 |
| chr1_-_22551594 | 0.00 |
ENSMUST00000239255.2
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0061181 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 1.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 | 0.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 1.8 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.3 | GO:0006855 | drug transmembrane transport(GO:0006855) organic cation transport(GO:0015695) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.4 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.8 | GO:0046332 | SMAD binding(GO:0046332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |