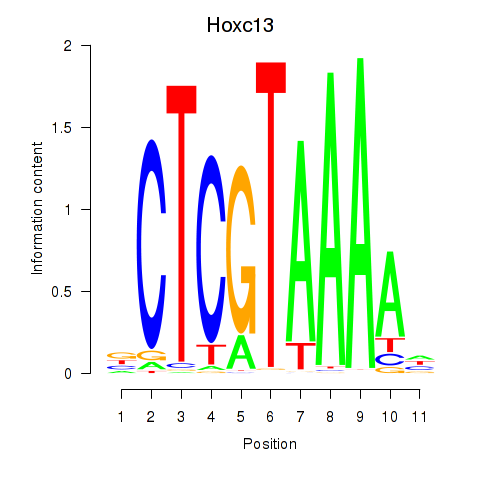
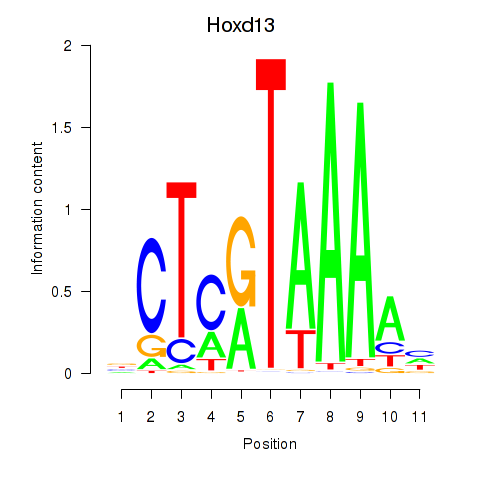
|
chr1_-_156767123
Show fit
|
0.26 |
ENSMUST00000189316.7
ENSMUST00000190648.7
ENSMUST00000172057.8
ENSMUST00000191605.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr6_+_108190050
Show fit
|
0.19 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1
|
|
chr6_+_108190163
Show fit
|
0.19 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1
|
|
chr8_-_70591799
Show fit
|
0.18 |
ENSMUST00000075724.9
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein
|
|
chr18_+_36431732
Show fit
|
0.16 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein
|
|
chr2_-_111815654
Show fit
|
0.16 |
ENSMUST00000214537.2
|
Olfr1309
|
olfactory receptor 1309
|
|
chr5_-_66238313
Show fit
|
0.15 |
ENSMUST00000202700.4
ENSMUST00000094757.9
ENSMUST00000113724.6
|
Rbm47
|
RNA binding motif protein 47
|
|
chr1_-_138775317
Show fit
|
0.14 |
ENSMUST00000093486.10
ENSMUST00000046870.13
|
Lhx9
|
LIM homeobox protein 9
|
|
chr9_+_113760376
Show fit
|
0.14 |
ENSMUST00000214095.2
ENSMUST00000116492.3
ENSMUST00000216558.2
|
Ubp1
|
upstream binding protein 1
|
|
chr1_+_59296065
Show fit
|
0.14 |
ENSMUST00000160662.8
ENSMUST00000114248.3
|
Cdk15
|
cyclin-dependent kinase 15
|
|
chr4_-_133329479
Show fit
|
0.12 |
ENSMUST00000057311.4
|
Sfn
|
stratifin
|
|
chr15_-_101336669
Show fit
|
0.12 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87
|
|
chr2_+_35022178
Show fit
|
0.12 |
ENSMUST00000113034.8
ENSMUST00000113037.10
ENSMUST00000150807.6
ENSMUST00000113033.6
|
Cntrl
|
centriolin
|
|
chr17_+_43878989
Show fit
|
0.12 |
ENSMUST00000167214.8
ENSMUST00000024706.12
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr19_-_7319211
Show fit
|
0.11 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2
|
|
chr7_-_73187369
Show fit
|
0.11 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2
|
|
chr13_-_19917092
Show fit
|
0.11 |
ENSMUST00000151029.3
|
Gpr141b
|
G protein-coupled receptor 141B
|
|
chr16_+_34815177
Show fit
|
0.10 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr19_-_7319157
Show fit
|
0.10 |
ENSMUST00000164205.8
ENSMUST00000165286.8
ENSMUST00000168324.2
ENSMUST00000032557.15
|
Mark2
|
MAP/microtubule affinity regulating kinase 2
|
|
chr19_+_18609343
Show fit
|
0.09 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1
|
|
chr1_-_156766957
Show fit
|
0.09 |
ENSMUST00000171292.8
ENSMUST00000063199.13
ENSMUST00000027886.14
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr11_+_77107006
Show fit
|
0.09 |
ENSMUST00000156488.8
ENSMUST00000037912.12
|
Ssh2
|
slingshot protein phosphatase 2
|
|
chr5_-_62923463
Show fit
|
0.08 |
ENSMUST00000076623.8
ENSMUST00000159470.3
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr6_-_112924205
Show fit
|
0.08 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr14_-_103584179
Show fit
|
0.08 |
ENSMUST00000159855.8
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase
|
|
chr3_+_79793237
Show fit
|
0.08 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B
|
|
chr4_-_148584906
Show fit
|
0.08 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7
|
|
chr10_-_33920300
Show fit
|
0.07 |
ENSMUST00000048052.7
|
Calhm4
|
calcium homeostasis modulator family member 4
|
|
chr1_-_156766381
Show fit
|
0.07 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr15_+_6552270
Show fit
|
0.07 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein
|
|
chr5_-_106606032
Show fit
|
0.07 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2
|
|
chrX_-_135769285
Show fit
|
0.06 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family
|
|
chr13_-_21586858
Show fit
|
0.06 |
ENSMUST00000117721.8
ENSMUST00000070785.16
ENSMUST00000116433.2
ENSMUST00000223831.2
ENSMUST00000116434.11
ENSMUST00000224820.2
|
Zkscan3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr10_+_56253418
Show fit
|
0.06 |
ENSMUST00000068581.9
ENSMUST00000217789.2
|
Gja1
|
gap junction protein, alpha 1
|
|
chr15_-_66158445
Show fit
|
0.06 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3
|
|
chr14_+_58308004
Show fit
|
0.06 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9
|
|
chr7_-_30312246
Show fit
|
0.06 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A
|
|
chr1_-_74163575
Show fit
|
0.06 |
ENSMUST00000169786.8
ENSMUST00000212888.2
ENSMUST00000191104.7
|
Tns1
|
tensin 1
|
|
chr17_+_46961250
Show fit
|
0.06 |
ENSMUST00000043464.14
|
Cul7
|
cullin 7
|
|
chr4_-_44710408
Show fit
|
0.06 |
ENSMUST00000134968.9
ENSMUST00000173821.8
ENSMUST00000174319.8
ENSMUST00000173733.8
ENSMUST00000172866.8
ENSMUST00000165417.9
ENSMUST00000107825.9
ENSMUST00000102932.10
ENSMUST00000107827.9
ENSMUST00000107826.9
ENSMUST00000014174.14
|
Pax5
|
paired box 5
|
|
chr9_+_38119661
Show fit
|
0.05 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893
|
|
chr3_+_107008867
Show fit
|
0.05 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2
|
|
chrX_-_7185424
Show fit
|
0.05 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5
|
|
chr11_+_60066681
Show fit
|
0.04 |
ENSMUST00000102688.2
|
Rai1
|
retinoic acid induced 1
|
|
chr9_+_32607301
Show fit
|
0.04 |
ENSMUST00000034534.13
ENSMUST00000050797.14
ENSMUST00000184887.2
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain
|
|
chr10_+_28544356
Show fit
|
0.04 |
ENSMUST00000060409.13
ENSMUST00000056097.11
ENSMUST00000105516.9
|
Themis
|
thymocyte selection associated
|
|
chr8_-_70591746
Show fit
|
0.04 |
ENSMUST00000212320.2
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein
|
|
chr2_+_74528071
Show fit
|
0.04 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9
|
|
chr1_+_72346572
Show fit
|
0.04 |
ENSMUST00000027379.10
|
Xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5
|
|
chrX_-_7185529
Show fit
|
0.03 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5
|
|
chr9_+_110948492
Show fit
|
0.03 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2
|
|
chr17_-_52133594
Show fit
|
0.03 |
ENSMUST00000129667.8
ENSMUST00000169480.8
ENSMUST00000148559.2
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr7_+_99876515
Show fit
|
0.03 |
ENSMUST00000084935.11
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr17_+_43879496
Show fit
|
0.03 |
ENSMUST00000169694.2
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr4_-_82768958
Show fit
|
0.03 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21
|
|
chr1_-_74639723
Show fit
|
0.03 |
ENSMUST00000127938.8
ENSMUST00000154874.8
|
Rnf25
|
ring finger protein 25
|
|
chr2_-_44912927
Show fit
|
0.03 |
ENSMUST00000202432.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr19_-_45619559
Show fit
|
0.03 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4
|
|
chr10_-_78688000
Show fit
|
0.03 |
ENSMUST00000205100.3
|
Olfr1356
|
olfactory receptor 1356
|
|
chr5_+_150119860
Show fit
|
0.03 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein
|
|
chr8_+_57004125
Show fit
|
0.03 |
ENSMUST00000110322.9
ENSMUST00000040218.13
ENSMUST00000210863.2
|
Fbxo8
|
F-box protein 8
|
|
chr2_-_73316053
Show fit
|
0.03 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_+_134042423
Show fit
|
0.02 |
ENSMUST00000105875.8
ENSMUST00000030638.7
|
Trim63
|
tripartite motif-containing 63
|
|
chr15_+_98972850
Show fit
|
0.02 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein
|
|
chr13_-_63712140
Show fit
|
0.02 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1
|
|
chr10_+_116137277
Show fit
|
0.02 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B
|
|
chr17_+_35658131
Show fit
|
0.02 |
ENSMUST00000071951.14
ENSMUST00000116598.10
ENSMUST00000078205.14
ENSMUST00000076256.8
|
H2-Q7
|
histocompatibility 2, Q region locus 7
|
|
chr1_+_85538554
Show fit
|
0.02 |
ENSMUST00000162925.2
|
Sp140
|
Sp140 nuclear body protein
|
|
chr6_-_71800788
Show fit
|
0.02 |
ENSMUST00000065103.4
|
Mrpl35
|
mitochondrial ribosomal protein L35
|
|
chr12_-_34578842
Show fit
|
0.02 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9
|
|
chr14_+_32043944
Show fit
|
0.02 |
ENSMUST00000022480.8
ENSMUST00000228529.2
|
Ogdhl
|
oxoglutarate dehydrogenase-like
|
|
chr8_-_37200051
Show fit
|
0.02 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1
|
|
chr13_+_35843816
Show fit
|
0.02 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like
|
|
chr16_-_57051727
Show fit
|
0.02 |
ENSMUST00000226586.2
|
Tbc1d23
|
TBC1 domain family, member 23
|
|
chr17_-_90395771
Show fit
|
0.02 |
ENSMUST00000197268.5
ENSMUST00000173917.8
|
Nrxn1
|
neurexin I
|
|
chr6_+_83063348
Show fit
|
0.01 |
ENSMUST00000041265.4
|
Lbx2
|
ladybird homeobox 2
|
|
chr4_-_58787716
Show fit
|
0.01 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267
|
|
chr1_-_156766351
Show fit
|
0.01 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr6_+_96092230
Show fit
|
0.01 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1
|
|
chr2_+_112096154
Show fit
|
0.01 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr4_+_21879660
Show fit
|
0.01 |
ENSMUST00000029909.3
|
Coq3
|
coenzyme Q3 methyltransferase
|
|
chr5_+_106024398
Show fit
|
0.01 |
ENSMUST00000150440.8
ENSMUST00000031227.11
|
Zfp326
|
zinc finger protein 326
|
|
chr11_-_5331734
Show fit
|
0.01 |
ENSMUST00000172492.8
|
Znrf3
|
zinc and ring finger 3
|
|
chr5_+_86952072
Show fit
|
0.01 |
ENSMUST00000119339.8
ENSMUST00000120498.8
|
Ythdc1
|
YTH domain containing 1
|
|
chr17_+_43879366
Show fit
|
0.01 |
ENSMUST00000167418.8
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr16_-_89305201
Show fit
|
0.01 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1
|
|
chr9_-_110775143
Show fit
|
0.01 |
ENSMUST00000199782.2
ENSMUST00000035075.13
|
Tdgf1
|
teratocarcinoma-derived growth factor 1
|
|
chr18_+_69478893
Show fit
|
0.01 |
ENSMUST00000202354.4
|
Tcf4
|
transcription factor 4
|
|
chr11_-_99920694
Show fit
|
0.01 |
ENSMUST00000073890.4
|
Krt33b
|
keratin 33B
|
|
chr8_-_25528972
Show fit
|
0.00 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4
|
|
chr18_-_46413886
Show fit
|
0.00 |
ENSMUST00000236999.2
|
Pggt1b
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr16_-_57051829
Show fit
|
0.00 |
ENSMUST00000023431.8
|
Tbc1d23
|
TBC1 domain family, member 23
|
|
chr11_-_23845207
Show fit
|
0.00 |
ENSMUST00000102863.3
ENSMUST00000020513.10
|
Papolg
|
poly(A) polymerase gamma
|
|
chr12_+_113038376
Show fit
|
0.00 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22
|
|
chr5_+_128677863
Show fit
|
0.00 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10
|
|
chr3_+_88528410
Show fit
|
0.00 |
ENSMUST00000029694.14
ENSMUST00000176804.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr7_-_30750828
Show fit
|
0.00 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7
|
|
chr18_+_36877709
Show fit
|
0.00 |
ENSMUST00000007042.6
ENSMUST00000237095.2
|
Ik
|
IK cytokine
|
|
chr7_-_102408573
Show fit
|
0.00 |
ENSMUST00000210453.3
ENSMUST00000232246.3
ENSMUST00000239110.2
ENSMUST00000060187.15
ENSMUST00000168007.3
|
Olfr560
Olfr78
|
olfactory receptor 560
olfactory receptor 78
|
|
chr7_-_30750856
Show fit
|
0.00 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7
|
|
chr10_+_87695886
Show fit
|
0.00 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1
|
|
chr15_-_101482320
Show fit
|
0.00 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75
|
|
chr12_+_113038397
Show fit
|
0.00 |
ENSMUST00000155492.2
|
Tex22
|
testis expressed gene 22
|