Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
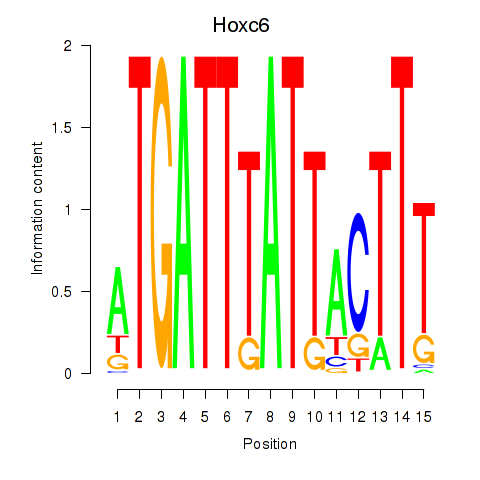
Results for Hoxc6
Z-value: 0.40
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSMUSG00000001661.6 | homeobox C6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc6 | mm39_v1_chr15_+_102917977_102918011 | -0.49 | 4.1e-01 | Click! |
Activity profile of Hoxc6 motif
Sorted Z-values of Hoxc6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_151143524 | 0.32 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr19_+_32597379 | 0.22 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr10_-_128462616 | 0.20 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr2_-_174188505 | 0.18 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chrM_+_14138 | 0.18 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.18 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_+_151143557 | 0.17 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr14_-_64654397 | 0.14 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chrM_+_9459 | 0.13 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_+_88030951 | 0.12 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr9_-_19088608 | 0.11 |
ENSMUST00000212127.3
|
Olfr839-ps1
|
olfactory receptor 839, pseudogene 1 |
| chr15_+_91557378 | 0.10 |
ENSMUST00000060642.7
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr19_+_11446716 | 0.10 |
ENSMUST00000165310.3
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr14_-_50586329 | 0.10 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr10_-_100425067 | 0.08 |
ENSMUST00000218821.2
ENSMUST00000054471.10 |
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr9_-_39161290 | 0.08 |
ENSMUST00000213472.2
|
Olfr1537
|
olfactory receptor 1537 |
| chr1_+_88062508 | 0.08 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr14_-_64654592 | 0.08 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr15_-_50752437 | 0.07 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr14_+_26722319 | 0.06 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr15_-_50753061 | 0.06 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_-_57340712 | 0.05 |
ENSMUST00000228156.2
ENSMUST00000227186.2 ENSMUST00000228294.2 ENSMUST00000228342.2 |
Vmn1r17
|
vomeronasal 1 receptor 17 |
| chr11_-_23845207 | 0.05 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr1_+_46464625 | 0.04 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C |
| chr7_+_73390026 | 0.04 |
ENSMUST00000107456.4
|
Fam174b
|
family with sequence similarity 174, member B |
| chr7_+_5023375 | 0.04 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr8_-_110688716 | 0.03 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr3_+_66892979 | 0.03 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr11_-_70560110 | 0.03 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr5_+_24618380 | 0.03 |
ENSMUST00000049346.10
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr14_-_52616625 | 0.03 |
ENSMUST00000214980.2
|
Olfr1512
|
olfactory receptor 1512 |
| chrM_+_8603 | 0.03 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr2_+_88470886 | 0.03 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chrX_-_142716200 | 0.03 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr5_-_129030367 | 0.03 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr10_+_112001581 | 0.02 |
ENSMUST00000170013.2
|
Caps2
|
calcyphosphine 2 |
| chrX_-_93256291 | 0.02 |
ENSMUST00000050328.15
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr5_-_23821523 | 0.02 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr3_-_151455514 | 0.02 |
ENSMUST00000029671.9
|
Ifi44
|
interferon-induced protein 44 |
| chr6_-_30693675 | 0.02 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr17_-_35334404 | 0.02 |
ENSMUST00000172854.2
ENSMUST00000062657.5 |
Ly6g5b
|
lymphocyte antigen 6 complex, locus G5B |
| chr17_+_3532455 | 0.02 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr13_+_93440572 | 0.02 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr17_+_38082190 | 0.02 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122 |
| chr6_-_123830422 | 0.02 |
ENSMUST00000162046.3
|
Vmn2r25
|
vomeronasal 2, receptor 25 |
| chr19_+_26725589 | 0.02 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_53745920 | 0.02 |
ENSMUST00000094964.7
|
Dnah7a
|
dynein, axonemal, heavy chain 7A |
| chr6_+_128376844 | 0.02 |
ENSMUST00000120405.4
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr8_-_84976330 | 0.01 |
ENSMUST00000019506.9
|
D8Ertd738e
|
DNA segment, Chr 8, ERATO Doi 738, expressed |
| chr4_+_147445744 | 0.01 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr7_-_109330915 | 0.01 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chr10_-_70491764 | 0.01 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr14_+_52680447 | 0.01 |
ENSMUST00000205900.3
ENSMUST00000215030.2 ENSMUST00000206100.2 ENSMUST00000206718.2 |
Olfr1509
|
olfactory receptor 1509 |
| chr11_+_109317046 | 0.01 |
ENSMUST00000238825.2
ENSMUST00000103061.3 |
Amz2
|
archaelysin family metallopeptidase 2 |
| chr4_-_52936281 | 0.01 |
ENSMUST00000217546.2
|
Olfr271-ps1
|
olfactory receptor 271, pseudogene 1 |
| chr9_+_40092216 | 0.01 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr9_-_76120939 | 0.01 |
ENSMUST00000074880.6
|
Gfral
|
GDNF family receptor alpha like |
| chr2_+_128704867 | 0.01 |
ENSMUST00000110324.8
|
Fbln7
|
fibulin 7 |
| chr4_+_33062999 | 0.01 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr2_+_65451100 | 0.01 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr4_-_147726953 | 0.01 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chr6_-_53955647 | 0.01 |
ENSMUST00000204674.3
ENSMUST00000166545.2 ENSMUST00000203101.3 |
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr17_+_38082519 | 0.01 |
ENSMUST00000119082.2
|
Olfr122
|
olfactory receptor 122 |
| chrX_-_142716085 | 0.01 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr10_+_19588318 | 0.01 |
ENSMUST00000020185.5
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr10_-_62285458 | 0.01 |
ENSMUST00000020273.16
|
Supv3l1
|
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr10_-_129591354 | 0.01 |
ENSMUST00000059038.3
|
Olfr807
|
olfactory receptor 807 |
| chr19_+_24853039 | 0.01 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr12_-_64521464 | 0.01 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr6_-_129894613 | 0.01 |
ENSMUST00000118060.8
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr10_+_5543769 | 0.01 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr9_-_39237341 | 0.01 |
ENSMUST00000216132.2
|
Olfr948
|
olfactory receptor 948 |
| chr6_+_36364990 | 0.01 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr10_-_107555840 | 0.01 |
ENSMUST00000050702.9
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chrX_-_166907286 | 0.01 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr5_+_17779273 | 0.01 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr2_-_89172273 | 0.01 |
ENSMUST00000215469.2
|
Olfr1233
|
olfactory receptor 1233 |
| chr2_-_51039112 | 0.01 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr3_-_113325938 | 0.01 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr10_+_23814470 | 0.01 |
ENSMUST00000079134.5
|
Taar2
|
trace amine-associated receptor 2 |
| chr7_+_108064533 | 0.01 |
ENSMUST00000217616.3
|
Olfr498
|
olfactory receptor 498 |
| chr2_-_111645069 | 0.01 |
ENSMUST00000099609.2
|
Olfr1303
|
olfactory receptor 1303 |
| chr19_+_39499288 | 0.00 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr11_-_94673526 | 0.00 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chrX_+_101846569 | 0.00 |
ENSMUST00000118218.8
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr2_-_85169639 | 0.00 |
ENSMUST00000111598.2
ENSMUST00000216347.2 |
Olfr987
|
olfactory receptor 987 |
| chr3_-_144738526 | 0.00 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr9_+_19351562 | 0.00 |
ENSMUST00000217273.2
|
Olfr849
|
olfactory receptor 849 |
| chr11_-_58624049 | 0.00 |
ENSMUST00000075607.5
|
Olfr317
|
olfactory receptor 317 |
| chr1_+_19279178 | 0.00 |
ENSMUST00000187754.7
|
Tfap2b
|
transcription factor AP-2 beta |
| chr14_-_52593865 | 0.00 |
ENSMUST00000215147.2
|
Olfr1513
|
olfactory receptor 1513 |
| chr9_+_65268304 | 0.00 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr2_-_85193571 | 0.00 |
ENSMUST00000215511.2
|
Olfr988
|
olfactory receptor 988 |
| chr7_+_107994038 | 0.00 |
ENSMUST00000210990.2
|
Olfr495
|
olfactory receptor 495 |
| chr3_+_133942244 | 0.00 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr11_+_73973733 | 0.00 |
ENSMUST00000178159.2
|
Zfp616
|
zinc finger protein 616 |
| chr2_-_87488801 | 0.00 |
ENSMUST00000099857.11
ENSMUST00000135875.4 |
Olfr1134
|
olfactory receptor 1134 |
| chr6_-_141719536 | 0.00 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr5_-_110194352 | 0.00 |
ENSMUST00000167969.2
|
Gm17655
|
predicted gene, 17655 |
| chr8_-_10026292 | 0.00 |
ENSMUST00000095476.6
|
Lig4
|
ligase IV, DNA, ATP-dependent |
| chr17_-_3746536 | 0.00 |
ENSMUST00000115800.2
|
Nox3
|
NADPH oxidase 3 |
| chr1_+_19279138 | 0.00 |
ENSMUST00000027059.11
|
Tfap2b
|
transcription factor AP-2 beta |
| chr12_+_29584560 | 0.00 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr11_+_58556387 | 0.00 |
ENSMUST00000072030.4
|
Olfr322
|
olfactory receptor 322 |
| chr8_-_55177510 | 0.00 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr6_-_3494587 | 0.00 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr2_-_86086061 | 0.00 |
ENSMUST00000099899.3
|
Olfr1049
|
olfactory receptor 1049 |
| chr2_-_111923044 | 0.00 |
ENSMUST00000099598.2
|
Olfr1314
|
olfactory receptor 1314 |
| chr5_-_129030111 | 0.00 |
ENSMUST00000196085.5
|
Rimbp2
|
RIMS binding protein 2 |
| chr6_-_57827328 | 0.00 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr10_-_108535970 | 0.00 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chrX_+_48552803 | 0.00 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr14_+_75479727 | 0.00 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr3_+_66893031 | 0.00 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_-_84464521 | 0.00 |
ENSMUST00000177040.2
|
Bnc2
|
basonuclin 2 |
| chr2_-_86048052 | 0.00 |
ENSMUST00000099902.2
|
Olfr1046
|
olfactory receptor 1046 |
| chr12_-_114226570 | 0.00 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chr8_+_46111778 | 0.00 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr4_-_36056726 | 0.00 |
ENSMUST00000108124.4
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr2_-_89172742 | 0.00 |
ENSMUST00000216561.2
|
Olfr1233
|
olfactory receptor 1233 |
| chr5_-_83502966 | 0.00 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr3_+_5283606 | 0.00 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr19_+_6414587 | 0.00 |
ENSMUST00000155973.2
|
Sf1
|
splicing factor 1 |
| chr19_+_11872971 | 0.00 |
ENSMUST00000072784.4
|
Olfr1420
|
olfactory receptor 1420 |
| chr7_-_115459082 | 0.00 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chrX_+_162945162 | 0.00 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.0 | 0.2 | GO:0033762 | response to glucagon(GO:0033762) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |