Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
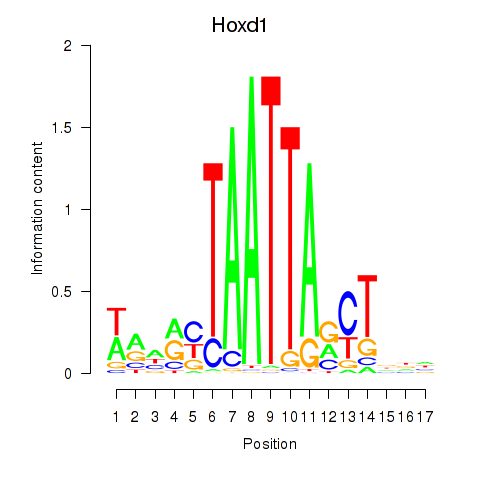
Results for Hoxd1
Z-value: 0.53
Transcription factors associated with Hoxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd1
|
ENSMUSG00000042448.6 | homeobox D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd1 | mm39_v1_chr2_+_74593324_74593324 | -0.49 | 4.0e-01 | Click! |
Activity profile of Hoxd1 motif
Sorted Z-values of Hoxd1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_9870 | 0.52 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_10167 | 0.37 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr15_-_8740218 | 0.30 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_129484070 | 0.28 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrM_+_8603 | 0.26 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chrM_+_7779 | 0.24 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr9_-_89586960 | 0.24 |
ENSMUST00000058488.9
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr1_+_87983099 | 0.21 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr7_-_24423715 | 0.20 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr1_+_87998487 | 0.20 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr11_+_58062467 | 0.20 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chrM_+_7758 | 0.17 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr8_-_41494890 | 0.16 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_32278245 | 0.15 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr1_-_65225572 | 0.15 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr7_-_44578834 | 0.14 |
ENSMUST00000107857.11
ENSMUST00000167930.8 ENSMUST00000085399.13 ENSMUST00000166972.9 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr14_+_69585036 | 0.14 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr6_+_134617903 | 0.14 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr17_-_36343573 | 0.14 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr12_+_10440755 | 0.14 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr10_+_128173603 | 0.14 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr1_-_65225617 | 0.13 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr2_+_69727563 | 0.13 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr7_+_99808526 | 0.13 |
ENSMUST00000207825.2
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr10_+_80003612 | 0.12 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr7_+_101545547 | 0.12 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr2_-_154916367 | 0.12 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr4_+_146695418 | 0.12 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr7_+_99808452 | 0.11 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr14_+_62529924 | 0.11 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr13_-_43634695 | 0.11 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr13_+_51799268 | 0.11 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr15_+_38662158 | 0.11 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr1_+_87983189 | 0.11 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr15_+_25774070 | 0.11 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr11_-_4045343 | 0.11 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr15_+_44482545 | 0.11 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr15_-_79658608 | 0.11 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr11_-_109502243 | 0.10 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr2_-_111650685 | 0.10 |
ENSMUST00000216114.3
|
Olfr1303
|
olfactory receptor 1303 |
| chr5_+_134961246 | 0.10 |
ENSMUST00000111218.8
ENSMUST00000136246.5 ENSMUST00000201847.3 |
Mettl27
|
methyltransferase like 27 |
| chr9_+_5298669 | 0.10 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr17_+_35150229 | 0.10 |
ENSMUST00000007253.6
|
Neu1
|
neuraminidase 1 |
| chr6_+_123239076 | 0.10 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr12_-_80690573 | 0.09 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr7_+_45271229 | 0.09 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr15_+_97990431 | 0.09 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr7_+_101546059 | 0.09 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr6_+_40619913 | 0.09 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr2_-_32277773 | 0.09 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr9_-_15212849 | 0.09 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_-_6217718 | 0.09 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
| chr11_+_109434519 | 0.09 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr5_+_102916637 | 0.09 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_134961205 | 0.09 |
ENSMUST00000047196.14
ENSMUST00000111221.9 ENSMUST00000111219.8 ENSMUST00000068617.12 |
Mettl27
|
methyltransferase like 27 |
| chr12_-_84664001 | 0.09 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr5_-_65855511 | 0.08 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr19_-_24178000 | 0.08 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr8_-_84976330 | 0.08 |
ENSMUST00000019506.9
|
D8Ertd738e
|
DNA segment, Chr 8, ERATO Doi 738, expressed |
| chr3_+_40755211 | 0.08 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr4_-_117539431 | 0.08 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr13_+_24985640 | 0.08 |
ENSMUST00000019276.12
|
BC005537
|
cDNA sequence BC005537 |
| chr9_+_119170486 | 0.08 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr8_-_106692668 | 0.07 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr5_-_3697806 | 0.07 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr19_-_41921676 | 0.07 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr2_-_20948230 | 0.07 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr10_-_128462616 | 0.07 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr11_-_30599510 | 0.07 |
ENSMUST00000074613.4
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr14_+_54701594 | 0.07 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr4_+_140427799 | 0.07 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr12_-_55061117 | 0.06 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr4_+_147216495 | 0.06 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr11_-_99121822 | 0.06 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr9_+_21634779 | 0.06 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr19_+_41921903 | 0.06 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr7_+_45364822 | 0.06 |
ENSMUST00000210640.2
|
Rpl18
|
ribosomal protein L18 |
| chr18_+_31742565 | 0.06 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr10_-_53252210 | 0.06 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr6_+_34006356 | 0.06 |
ENSMUST00000228187.2
ENSMUST00000070189.10 ENSMUST00000101564.9 |
Lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr19_-_53932867 | 0.06 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr9_-_45115381 | 0.06 |
ENSMUST00000034599.15
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr5_+_138185747 | 0.06 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr4_-_132073048 | 0.05 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr11_-_45846291 | 0.05 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr12_+_72488625 | 0.05 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr13_+_24986003 | 0.05 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr3_+_32490300 | 0.05 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr4_-_132072988 | 0.05 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr11_-_115310743 | 0.05 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr15_-_79658584 | 0.05 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr11_-_45845992 | 0.05 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr6_-_69282389 | 0.05 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr8_+_46463633 | 0.05 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr18_+_37898633 | 0.05 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr2_-_168609110 | 0.05 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr2_+_32496990 | 0.05 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr4_+_145397238 | 0.04 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr19_-_32173824 | 0.04 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr1_-_4430481 | 0.04 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr4_+_147576874 | 0.04 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr19_-_53932581 | 0.04 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr4_+_146586445 | 0.04 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr8_-_95929420 | 0.04 |
ENSMUST00000212842.2
|
Kifc3
|
kinesin family member C3 |
| chr15_+_44482667 | 0.04 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr4_+_134658209 | 0.04 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chrX_-_98514278 | 0.04 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr2_-_150097511 | 0.04 |
ENSMUST00000063463.6
|
Gm21994
|
predicted gene 21994 |
| chr10_-_112764879 | 0.04 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr17_+_38104420 | 0.04 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr2_-_69542805 | 0.03 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr4_+_146599377 | 0.03 |
ENSMUST00000140089.7
ENSMUST00000179175.3 |
Zfp981
|
zinc finger protein 981 |
| chr3_-_120965327 | 0.03 |
ENSMUST00000170781.2
ENSMUST00000039761.12 ENSMUST00000106467.8 ENSMUST00000106466.10 ENSMUST00000164925.9 |
Rwdd3
|
RWD domain containing 3 |
| chr11_-_99265721 | 0.03 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr4_-_14621805 | 0.03 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_-_172212426 | 0.03 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr5_-_138185438 | 0.03 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr13_-_103042554 | 0.03 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_32496957 | 0.03 |
ENSMUST00000113290.8
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr17_-_56343625 | 0.03 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr13_-_74956030 | 0.03 |
ENSMUST00000065629.6
|
Cast
|
calpastatin |
| chr10_-_129107354 | 0.03 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr13_-_74956640 | 0.03 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr17_+_38104635 | 0.03 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr1_+_161796854 | 0.03 |
ENSMUST00000160881.2
ENSMUST00000159648.2 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr18_+_24737009 | 0.03 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr11_+_60428788 | 0.03 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr10_+_94411119 | 0.03 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr2_+_85809620 | 0.03 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr16_-_19341016 | 0.03 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr12_+_80690985 | 0.03 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr2_-_86109346 | 0.03 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr6_-_130314465 | 0.03 |
ENSMUST00000088017.5
ENSMUST00000111998.9 |
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3 |
| chr8_-_46542730 | 0.03 |
ENSMUST00000144244.2
|
Snx25
|
sorting nexin 25 |
| chr19_+_8779903 | 0.03 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr7_+_107411470 | 0.03 |
ENSMUST00000208563.3
ENSMUST00000214253.2 |
Olfr467
|
olfactory receptor 467 |
| chr2_-_132089667 | 0.03 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr19_+_13316226 | 0.03 |
ENSMUST00000207124.3
|
Olfr1466
|
olfactory receptor 1466 |
| chr5_-_86437119 | 0.03 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr2_-_168608949 | 0.03 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr2_-_17465410 | 0.03 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chrX_-_158921370 | 0.03 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr17_-_56343531 | 0.03 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr9_+_53678801 | 0.03 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr1_-_4479445 | 0.02 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr4_+_145241454 | 0.02 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr6_-_130106861 | 0.02 |
ENSMUST00000014476.6
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr3_+_55689921 | 0.02 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr2_+_85612365 | 0.02 |
ENSMUST00000215945.2
|
Olfr1015
|
olfactory receptor 1015 |
| chr6_-_69162381 | 0.02 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr14_-_70666513 | 0.02 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr8_-_62576140 | 0.02 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr19_-_7943365 | 0.02 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr10_+_129493563 | 0.02 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr3_+_104688363 | 0.02 |
ENSMUST00000002298.7
|
Ppm1j
|
protein phosphatase 1J |
| chrX_+_56257374 | 0.02 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr11_-_107228382 | 0.02 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr2_-_27365633 | 0.02 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr18_+_37488174 | 0.02 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr6_-_131667026 | 0.02 |
ENSMUST00000080619.3
|
Tas2r114
|
taste receptor, type 2, member 114 |
| chr5_+_90708962 | 0.02 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr2_-_85992176 | 0.02 |
ENSMUST00000099908.6
ENSMUST00000215624.2 |
Olfr1042
|
olfactory receptor 1042 |
| chr2_+_121786892 | 0.02 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chrX_-_142716085 | 0.02 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr19_-_56378309 | 0.02 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr1_+_190660689 | 0.02 |
ENSMUST00000066632.14
ENSMUST00000110899.7 |
Angel2
|
angel homolog 2 |
| chr14_+_65504067 | 0.02 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr4_+_146033882 | 0.02 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr14_+_25980463 | 0.02 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr12_+_112727089 | 0.02 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr13_-_21847556 | 0.02 |
ENSMUST00000214321.2
|
Olfr1361
|
olfactory receptor 1361 |
| chr2_+_85868891 | 0.02 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr8_-_79975199 | 0.02 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr9_+_38738911 | 0.02 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr14_-_54651442 | 0.02 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr3_-_87081939 | 0.02 |
ENSMUST00000159976.8
ENSMUST00000107618.9 |
Kirrel
|
kirre like nephrin family adhesion molecule 1 |
| chr2_+_88505972 | 0.02 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr11_-_117716918 | 0.02 |
ENSMUST00000026661.4
|
Tk1
|
thymidine kinase 1 |
| chr2_-_89127612 | 0.02 |
ENSMUST00000099787.2
|
Olfr1230
|
olfactory receptor 1230 |
| chr13_-_57043550 | 0.02 |
ENSMUST00000022023.13
ENSMUST00000174457.9 ENSMUST00000109871.8 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr18_-_3281089 | 0.02 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr2_-_86088672 | 0.02 |
ENSMUST00000216185.3
|
Olfr1049
|
olfactory receptor 1049 |
| chr13_-_21330037 | 0.02 |
ENSMUST00000216039.3
|
Olfr1368
|
olfactory receptor 1368 |
| chr2_-_89159994 | 0.02 |
ENSMUST00000213860.2
ENSMUST00000215679.2 |
Olfr1232
|
olfactory receptor 1232 |
| chr6_-_124756645 | 0.02 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr2_+_65451100 | 0.02 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr5_-_138185686 | 0.02 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr1_-_171854818 | 0.02 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr14_-_51308899 | 0.02 |
ENSMUST00000048478.6
|
Olfr750
|
olfactory receptor 750 |
| chr2_-_87570322 | 0.02 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr2_-_164427367 | 0.02 |
ENSMUST00000109342.2
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr7_+_20896110 | 0.02 |
ENSMUST00000166948.2
|
Vmn1r123
|
vomeronasal 1 receptor 123 |
| chr10_+_57527084 | 0.02 |
ENSMUST00000175852.2
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr3_+_60503051 | 0.02 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr4_+_109092459 | 0.02 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr14_-_52341472 | 0.02 |
ENSMUST00000111610.12
ENSMUST00000164655.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr7_-_103094646 | 0.02 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr6_-_131655849 | 0.02 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr19_-_40371016 | 0.02 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr7_-_19939532 | 0.02 |
ENSMUST00000169774.2
|
Vmn1r241
|
vomeronasal 1 receptor 241 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:2000566 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |