Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxd10
Z-value: 0.27
Transcription factors associated with Hoxd10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd10
|
ENSMUSG00000050368.5 | homeobox D10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd10 | mm39_v1_chr2_+_74522258_74522284 | 0.59 | 2.9e-01 | Click! |
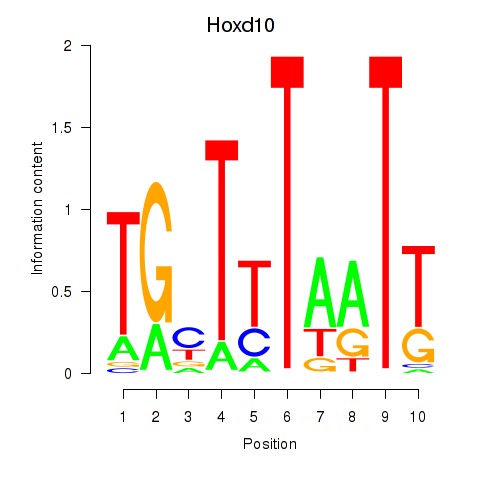
Activity profile of Hoxd10 motif
Sorted Z-values of Hoxd10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_49735938 | 0.14 |
ENSMUST00000221170.2
|
Omd
|
osteomodulin |
| chr3_+_106393348 | 0.10 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr10_+_21171878 | 0.08 |
ENSMUST00000020153.10
ENSMUST00000218032.2 ENSMUST00000061324.6 ENSMUST00000219915.2 ENSMUST00000092674.14 ENSMUST00000218714.2 |
Hbs1l
|
Hbs1-like (S. cerevisiae) |
| chr7_+_81512421 | 0.08 |
ENSMUST00000119543.2
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr19_+_45433899 | 0.07 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr10_+_116013256 | 0.07 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr14_-_70945434 | 0.06 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr5_-_65090104 | 0.06 |
ENSMUST00000059349.6
|
Tlr1
|
toll-like receptor 1 |
| chr14_+_79753055 | 0.05 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr5_+_150218847 | 0.05 |
ENSMUST00000239118.2
|
Fry
|
FRY microtubule binding protein |
| chr4_+_102112189 | 0.05 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_-_14873406 | 0.04 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr1_-_149798090 | 0.04 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr16_-_37681508 | 0.04 |
ENSMUST00000205931.2
|
Gm36028
|
predicted gene, 36028 |
| chr14_-_50479161 | 0.04 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr15_+_57775595 | 0.04 |
ENSMUST00000022992.13
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr10_-_30494333 | 0.04 |
ENSMUST00000019925.7
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr10_+_128626772 | 0.03 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr12_+_117652526 | 0.03 |
ENSMUST00000222185.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_+_33226020 | 0.03 |
ENSMUST00000052266.15
ENSMUST00000090381.11 ENSMUST00000115080.2 |
Exoc4
|
exocyst complex component 4 |
| chr10_-_12490424 | 0.03 |
ENSMUST00000217994.2
|
Utrn
|
utrophin |
| chr13_+_38529062 | 0.03 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr19_-_24202344 | 0.03 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr14_-_54655079 | 0.03 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr9_-_45739886 | 0.03 |
ENSMUST00000214868.2
ENSMUST00000117194.8 |
Cep164
|
centrosomal protein 164 |
| chr3_-_130524024 | 0.03 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr9_-_56151334 | 0.02 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr4_+_100336003 | 0.02 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr11_-_99134885 | 0.02 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr10_-_13199971 | 0.02 |
ENSMUST00000105543.9
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr9_+_110948492 | 0.02 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr7_+_51530060 | 0.02 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr2_+_120235239 | 0.01 |
ENSMUST00000135074.2
|
Ganc
|
glucosidase, alpha; neutral C |
| chr4_-_58787716 | 0.01 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267 |
| chrX_+_65696608 | 0.01 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr12_-_4891435 | 0.01 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr7_-_73191484 | 0.01 |
ENSMUST00000197642.2
ENSMUST00000026895.14 ENSMUST00000169922.9 |
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr2_+_109522781 | 0.01 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr10_-_30494431 | 0.01 |
ENSMUST00000161074.8
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr6_-_28397997 | 0.01 |
ENSMUST00000035930.11
|
Zfp800
|
zinc finger protein 800 |
| chr17_+_78815493 | 0.01 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr13_+_49736219 | 0.01 |
ENSMUST00000065494.8
|
Omd
|
osteomodulin |
| chrX_-_48823936 | 0.01 |
ENSMUST00000215373.3
|
Olfr1321
|
olfactory receptor 1321 |
| chr15_-_103123711 | 0.01 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr18_+_5035072 | 0.01 |
ENSMUST00000210707.2
|
Svil
|
supervillin |
| chr7_+_43339842 | 0.01 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr4_-_116228921 | 0.01 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr3_-_75177378 | 0.01 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr19_+_20470056 | 0.01 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr19_+_12972378 | 0.01 |
ENSMUST00000207997.3
|
Olfr1451
|
olfactory receptor 1451 |
| chr1_-_132318039 | 0.00 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr4_+_43401232 | 0.00 |
ENSMUST00000125399.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr6_-_23655130 | 0.00 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr6_-_129752812 | 0.00 |
ENSMUST00000095409.3
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr2_-_76045696 | 0.00 |
ENSMUST00000144892.2
|
Pde11a
|
phosphodiesterase 11A |
| chr5_-_73787646 | 0.00 |
ENSMUST00000152215.3
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr6_+_70663930 | 0.00 |
ENSMUST00000103402.2
|
Igkv3-3
|
immunoglobulin kappa variable 3-3 |
| chr15_+_44059531 | 0.00 |
ENSMUST00000038856.14
ENSMUST00000110289.4 |
Trhr
|
thyrotropin releasing hormone receptor |
| chr10_-_107747995 | 0.00 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr9_+_38755745 | 0.00 |
ENSMUST00000217350.2
|
Olfr924
|
olfactory receptor 924 |
| chr2_+_87640166 | 0.00 |
ENSMUST00000090707.2
ENSMUST00000079711.3 |
Olfr1145
|
olfactory receptor 1145 |
| chr19_+_34078333 | 0.00 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr17_-_66901568 | 0.00 |
ENSMUST00000024914.4
|
Themis3
|
thymocyte selection associated family member 3 |
| chr1_+_139429430 | 0.00 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr9_+_38120508 | 0.00 |
ENSMUST00000212815.2
|
Olfr893
|
olfactory receptor 893 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |