Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
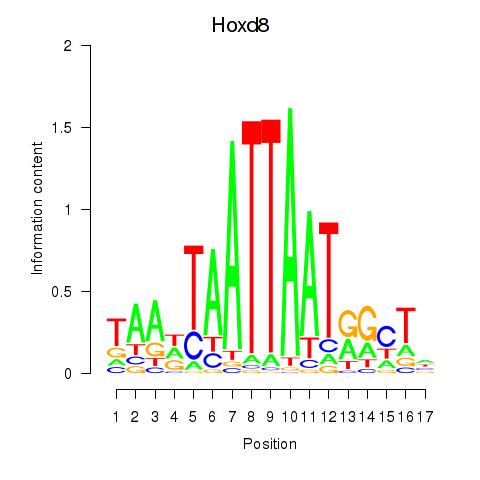
Results for Hoxd8
Z-value: 0.33
Transcription factors associated with Hoxd8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd8
|
ENSMUSG00000027102.5 | homeobox D8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd8 | mm39_v1_chr2_+_74534959_74534992 | -0.48 | 4.1e-01 | Click! |
Activity profile of Hoxd8 motif
Sorted Z-values of Hoxd8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_65855511 | 0.20 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr10_-_126866682 | 0.20 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr14_+_69585036 | 0.18 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr2_+_36120438 | 0.18 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_-_19319155 | 0.16 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr9_-_55419442 | 0.15 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr6_+_134617903 | 0.14 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr9_+_21634779 | 0.14 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chrM_+_7758 | 0.13 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr18_+_11766333 | 0.13 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr12_+_87490666 | 0.12 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr12_-_84664001 | 0.12 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr14_-_86986541 | 0.12 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr18_-_43610829 | 0.11 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr15_+_65682066 | 0.11 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr17_+_46807637 | 0.11 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr14_+_55797443 | 0.11 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr8_+_34089597 | 0.10 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| chrX_-_158921370 | 0.10 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr14_+_26722319 | 0.09 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_-_126342551 | 0.09 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr17_-_56343531 | 0.09 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr6_+_34361153 | 0.09 |
ENSMUST00000038383.14
ENSMUST00000115051.8 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr1_-_171854818 | 0.09 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr12_-_87490580 | 0.09 |
ENSMUST00000162961.8
ENSMUST00000185301.2 |
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr8_-_85389470 | 0.09 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr1_-_149836974 | 0.08 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr2_+_83554741 | 0.08 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr11_+_109434519 | 0.08 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr1_+_9618173 | 0.07 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr10_+_79304565 | 0.07 |
ENSMUST00000233065.2
ENSMUST00000167976.4 |
Vmn2r83
|
vomeronasal 2, receptor 83 |
| chr17_+_74796473 | 0.07 |
ENSMUST00000024873.7
|
Yipf4
|
Yip1 domain family, member 4 |
| chr7_-_84661476 | 0.07 |
ENSMUST00000124773.3
ENSMUST00000233725.2 ENSMUST00000233739.2 ENSMUST00000232837.2 |
Vmn2r66
|
vomeronasal 2, receptor 66 |
| chr10_-_80754016 | 0.07 |
ENSMUST00000057623.14
ENSMUST00000179022.8 |
Lmnb2
|
lamin B2 |
| chr10_+_58230203 | 0.07 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_-_15212849 | 0.07 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr19_-_38031774 | 0.06 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr1_+_58841808 | 0.06 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr9_+_111011388 | 0.06 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr10_-_126866658 | 0.06 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr17_-_8366536 | 0.06 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chr10_+_116013256 | 0.06 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr13_-_63036096 | 0.06 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr15_+_99192968 | 0.06 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr16_+_33614715 | 0.05 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr8_+_40807344 | 0.05 |
ENSMUST00000136835.2
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chrX_-_73436609 | 0.05 |
ENSMUST00000015427.13
|
Fam3a
|
family with sequence similarity 3, member A |
| chr12_-_25147139 | 0.05 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chrX_+_20529137 | 0.05 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr7_+_140343652 | 0.04 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr7_-_99629637 | 0.04 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr1_+_87983099 | 0.04 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_+_72714156 | 0.04 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr9_+_53678801 | 0.04 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr17_+_79919267 | 0.04 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr18_+_31742565 | 0.04 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr10_-_25076008 | 0.04 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr4_-_108158242 | 0.04 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chrM_+_7779 | 0.04 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr14_+_65612788 | 0.03 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr10_+_129219952 | 0.03 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr1_+_87983189 | 0.03 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chrX_-_73436293 | 0.03 |
ENSMUST00000114138.8
|
Fam3a
|
family with sequence similarity 3, member A |
| chr14_+_55797468 | 0.03 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr9_-_82856321 | 0.03 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr13_+_38388904 | 0.03 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr5_-_73349191 | 0.03 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr9_+_111011327 | 0.03 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_89642395 | 0.03 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr19_+_29902506 | 0.03 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr4_+_147056433 | 0.03 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr3_-_14843512 | 0.03 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr8_-_117809188 | 0.03 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr3_-_19319123 | 0.03 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr10_+_116013122 | 0.02 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_-_99302205 | 0.02 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr2_+_110551685 | 0.02 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr2_+_110427643 | 0.02 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr19_-_18978463 | 0.02 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr2_+_110551927 | 0.02 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr5_-_38649291 | 0.02 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr14_-_20443676 | 0.02 |
ENSMUST00000061444.5
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr3_+_88624194 | 0.02 |
ENSMUST00000172252.2
|
Rit1
|
Ras-like without CAAX 1 |
| chr11_+_99748741 | 0.02 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr13_+_24023428 | 0.02 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr17_-_56343625 | 0.02 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr16_+_18247666 | 0.02 |
ENSMUST00000144233.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr3_+_20011405 | 0.02 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr2_+_87686206 | 0.02 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr9_-_57065572 | 0.01 |
ENSMUST00000065358.9
|
Commd4
|
COMM domain containing 4 |
| chr4_+_109092459 | 0.01 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr15_+_57775595 | 0.01 |
ENSMUST00000022992.13
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr2_+_151336095 | 0.01 |
ENSMUST00000089140.13
|
Nsfl1c
|
NSFL1 (p97) cofactor (p47) |
| chr4_+_109092610 | 0.01 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr3_-_144555062 | 0.01 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr2_-_86640362 | 0.01 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr3_+_88624145 | 0.01 |
ENSMUST00000029692.15
ENSMUST00000171645.8 |
Rit1
|
Ras-like without CAAX 1 |
| chr7_+_130633776 | 0.01 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr1_-_173703424 | 0.01 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr3_+_75982890 | 0.01 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr5_-_121665249 | 0.01 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr5_-_23889607 | 0.01 |
ENSMUST00000197985.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_+_103197281 | 0.01 |
ENSMUST00000214173.2
|
Olfr613
|
olfactory receptor 613 |
| chr11_+_73489420 | 0.01 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr3_-_72875187 | 0.01 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr10_-_130020042 | 0.01 |
ENSMUST00000216661.2
ENSMUST00000203720.3 |
Olfr826
|
olfactory receptor 826 |
| chr3_+_142300601 | 0.01 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr2_-_88854443 | 0.01 |
ENSMUST00000099799.5
|
Olfr1217
|
olfactory receptor 1217 |
| chr14_+_96118660 | 0.01 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr3_-_75177378 | 0.01 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr5_+_88523967 | 0.01 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr7_+_44711853 | 0.01 |
ENSMUST00000107829.9
ENSMUST00000003513.11 ENSMUST00000211465.2 ENSMUST00000210088.2 ENSMUST00000210520.2 |
Nosip
|
nitric oxide synthase interacting protein |
| chrX_+_9751861 | 0.01 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr10_-_103072141 | 0.01 |
ENSMUST00000123364.2
ENSMUST00000166240.8 ENSMUST00000020043.12 |
Lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr6_+_41331039 | 0.01 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr8_+_46463633 | 0.01 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr4_-_96673423 | 0.01 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr1_+_106866678 | 0.01 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr3_+_94280101 | 0.01 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr10_+_129493563 | 0.01 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr5_+_29400981 | 0.01 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr6_+_65502292 | 0.01 |
ENSMUST00000212402.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr2_-_66240408 | 0.01 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr11_+_87457479 | 0.01 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chr8_-_65489834 | 0.01 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr4_-_3185358 | 0.01 |
ENSMUST00000105159.5
|
Vmn1r3
|
vomeronasal 1 receptor 3 |
| chr15_-_96929086 | 0.01 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr6_-_141719536 | 0.01 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr6_-_3399451 | 0.01 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr2_+_87248394 | 0.01 |
ENSMUST00000054974.2
|
Olfr1123
|
olfactory receptor 1123 |
| chr6_+_65502344 | 0.01 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr7_+_20521210 | 0.01 |
ENSMUST00000173723.2
ENSMUST00000168794.2 |
Vmn1r113
|
vomeronasal 1 receptor 113 |
| chr2_+_85868891 | 0.01 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr2_-_86276348 | 0.01 |
ENSMUST00000216165.2
|
Olfr1065
|
olfactory receptor 1065 |
| chr3_-_33136153 | 0.01 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr2_-_86372824 | 0.01 |
ENSMUST00000216480.2
ENSMUST00000111582.3 |
Olfr1079
|
olfactory receptor 1079 |
| chr1_+_88234454 | 0.01 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr2_-_111452417 | 0.01 |
ENSMUST00000099612.2
|
Olfr1297
|
olfactory receptor 1297 |
| chr5_-_86780277 | 0.01 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr2_+_110551976 | 0.00 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr19_-_39637489 | 0.00 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr4_+_52596266 | 0.00 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr14_+_50645025 | 0.00 |
ENSMUST00000214853.2
ENSMUST00000214320.2 |
Olfr738
|
olfactory receptor 738 |
| chr2_+_87362140 | 0.00 |
ENSMUST00000217113.2
|
Olfr153
|
olfactory receptor 153 |
| chr19_+_13316226 | 0.00 |
ENSMUST00000207124.3
|
Olfr1466
|
olfactory receptor 1466 |
| chr16_-_19443851 | 0.00 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chrY_-_40270796 | 0.00 |
ENSMUST00000177713.2
|
Gm21865
|
predicted gene, 21865 |
| chr7_+_104583447 | 0.00 |
ENSMUST00000214260.2
ENSMUST00000210138.3 |
Olfr669
|
olfactory receptor 669 |
| chr2_+_85836999 | 0.00 |
ENSMUST00000079298.5
|
Olfr1032
|
olfactory receptor 1032 |
| chr6_+_30610973 | 0.00 |
ENSMUST00000062758.11
|
Cpa5
|
carboxypeptidase A5 |
| chr9_-_39427105 | 0.00 |
ENSMUST00000216177.2
|
Olfr957
|
olfactory receptor 957 |
| chr6_-_30936013 | 0.00 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr13_+_24023386 | 0.00 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr1_-_126758369 | 0.00 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr9_+_18855946 | 0.00 |
ENSMUST00000060601.5
|
Olfr832
|
olfactory receptor 832 |
| chr19_-_46661321 | 0.00 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr19_-_46661501 | 0.00 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr2_-_88110219 | 0.00 |
ENSMUST00000099830.3
|
Olfr1173
|
olfactory receptor 1173 |
| chr15_-_101422054 | 0.00 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr2_-_88905433 | 0.00 |
ENSMUST00000099797.2
|
Olfr1219
|
olfactory receptor 1219 |
| chr13_+_22474090 | 0.00 |
ENSMUST00000228382.2
ENSMUST00000228557.2 ENSMUST00000226245.2 ENSMUST00000227516.2 |
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chrY_+_52778041 | 0.00 |
ENSMUST00000178673.2
|
Gm21258
|
predicted gene, 21258 |
| chr6_+_30611028 | 0.00 |
ENSMUST00000115138.8
|
Cpa5
|
carboxypeptidase A5 |
| chrY_-_35085749 | 0.00 |
ENSMUST00000180170.2
|
Gm20855
|
predicted gene, 20855 |
| chrY_-_35875442 | 0.00 |
ENSMUST00000180332.2
|
Gm20896
|
predicted gene, 20896 |
| chr19_-_55229668 | 0.00 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr5_-_23889591 | 0.00 |
ENSMUST00000198549.2
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chrY_+_80146479 | 0.00 |
ENSMUST00000179811.2
|
Gm21760
|
predicted gene, 21760 |
| chr11_-_59466995 | 0.00 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr3_-_96104886 | 0.00 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chrY_+_84109980 | 0.00 |
ENSMUST00000177775.2
|
Gm21095
|
predicted gene, 21095 |
| chrY_+_79332266 | 0.00 |
ENSMUST00000178063.2
|
Gm20916
|
predicted gene, 20916 |
| chr16_-_19425453 | 0.00 |
ENSMUST00000078603.3
ENSMUST00000218837.2 |
Olfr170
|
olfactory receptor 170 |
| chr14_-_52648384 | 0.00 |
ENSMUST00000079459.4
|
Olfr1510
|
olfactory receptor 1510 |
| chr9_+_38240356 | 0.00 |
ENSMUST00000214155.2
ENSMUST00000212354.3 |
Olfr25
|
olfactory receptor 25 |
| chr2_+_87820457 | 0.00 |
ENSMUST00000102622.2
|
Olfr1158
|
olfactory receptor 1158 |
| chr4_+_88768324 | 0.00 |
ENSMUST00000094972.2
|
Ifna1
|
interferon alpha 1 |
| chr7_-_19856504 | 0.00 |
ENSMUST00000079099.2
|
Vmn1r240
|
vomeronasal 1 receptor 240 |
| chr2_+_83554868 | 0.00 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr2_-_140513382 | 0.00 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr16_-_75382905 | 0.00 |
ENSMUST00000062721.11
|
Lipi
|
lipase, member I |
| chrY_+_55211732 | 0.00 |
ENSMUST00000180249.2
|
Gm20931
|
predicted gene, 20931 |
| chr7_-_20279682 | 0.00 |
ENSMUST00000173886.2
|
Vmn1r245
|
vomeronasal 1 receptor 245 |
| chr6_+_24733239 | 0.00 |
ENSMUST00000031690.6
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chrX_-_105884178 | 0.00 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chrY_+_65387652 | 0.00 |
ENSMUST00000178198.2
|
Gm20736
|
predicted gene, 20736 |
| chr3_-_69767208 | 0.00 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr16_+_14407073 | 0.00 |
ENSMUST00000100165.4
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr5_-_103777145 | 0.00 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr7_-_21543692 | 0.00 |
ENSMUST00000105195.2
|
Vmn1r249
|
vomeronasal 1 receptor 249 |
| chr16_-_59107071 | 0.00 |
ENSMUST00000201687.3
ENSMUST00000217485.2 |
Olfr202
|
olfactory receptor 202 |
| chr2_-_87958616 | 0.00 |
ENSMUST00000217575.3
|
Olfr1166
|
olfactory receptor 1166 |
| chr5_-_66672158 | 0.00 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr2_-_140513320 | 0.00 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr2_-_88928307 | 0.00 |
ENSMUST00000099789.5
|
Olfr1220
|
olfactory receptor 1220 |
| chr5_+_87148697 | 0.00 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr2_-_86488800 | 0.00 |
ENSMUST00000099879.2
|
Olfr1085
|
olfactory receptor 1085 |
| chr1_+_88093726 | 0.00 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr11_-_99979052 | 0.00 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr2_+_88012224 | 0.00 |
ENSMUST00000215996.2
|
Olfr1168
|
olfactory receptor 1168 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0061346 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.2 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |