Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxd9
Z-value: 0.94
Transcription factors associated with Hoxd9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd9
|
ENSMUSG00000043342.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd9 | mm39_v1_chr2_+_74528071_74528097 | -0.82 | 8.8e-02 | Click! |
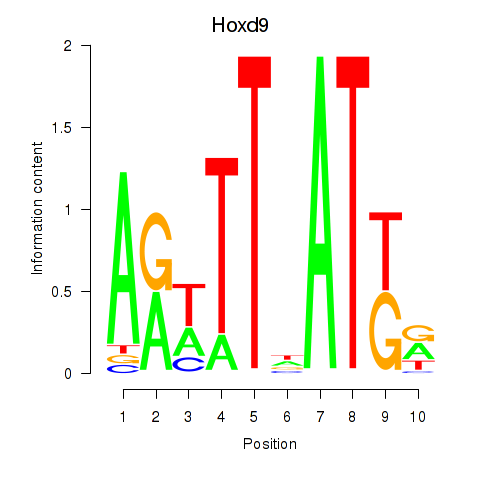
Activity profile of Hoxd9 motif
Sorted Z-values of Hoxd9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_35587888 | 0.72 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr14_+_66043281 | 0.63 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr14_+_74973081 | 0.59 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chrX_-_133442596 | 0.57 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chrX_+_55493325 | 0.54 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr2_-_151586063 | 0.48 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr9_-_64080161 | 0.46 |
ENSMUST00000176299.8
ENSMUST00000130127.8 ENSMUST00000176794.8 ENSMUST00000177045.8 |
Zwilch
|
zwilch kinetochore protein |
| chr5_-_21087023 | 0.44 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr12_+_117807224 | 0.44 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chrX_+_41241049 | 0.41 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr14_-_54855446 | 0.39 |
ENSMUST00000227257.2
ENSMUST00000022803.6 |
Psmb5
|
proteasome (prosome, macropain) subunit, beta type 5 |
| chr11_+_23234644 | 0.39 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr3_-_115923098 | 0.38 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr1_-_154975376 | 0.38 |
ENSMUST00000055322.6
|
Ier5
|
immediate early response 5 |
| chr6_+_18848600 | 0.35 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr16_+_4559426 | 0.33 |
ENSMUST00000120232.8
|
Hmox2
|
heme oxygenase 2 |
| chr5_-_113458563 | 0.33 |
ENSMUST00000154248.8
ENSMUST00000112325.8 ENSMUST00000048112.13 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr14_+_43951187 | 0.32 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr16_-_65359406 | 0.32 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr8_+_85583611 | 0.31 |
ENSMUST00000003906.13
ENSMUST00000109754.2 |
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr5_-_69748126 | 0.30 |
ENSMUST00000166298.8
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr7_-_84328553 | 0.30 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_-_97689263 | 0.29 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr10_-_93727003 | 0.29 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr9_-_60594742 | 0.27 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr3_+_5815863 | 0.26 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr15_-_98832403 | 0.26 |
ENSMUST00000077577.8
|
Tuba1b
|
tubulin, alpha 1B |
| chr2_-_84255602 | 0.25 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr1_-_138103021 | 0.25 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_+_57679621 | 0.25 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr11_-_80030735 | 0.23 |
ENSMUST00000136996.2
|
Tefm
|
transcription elongation factor, mitochondrial |
| chrX_-_10083157 | 0.22 |
ENSMUST00000072393.9
ENSMUST00000044598.13 ENSMUST00000073392.11 ENSMUST00000115533.8 ENSMUST00000115532.2 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr1_-_138102972 | 0.22 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_32490300 | 0.22 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr7_+_127399776 | 0.22 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_+_35808269 | 0.22 |
ENSMUST00000029257.15
|
Atp11b
|
ATPase, class VI, type 11B |
| chr18_+_31922173 | 0.21 |
ENSMUST00000025106.5
ENSMUST00000234146.2 |
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr3_+_103078971 | 0.19 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr5_+_136982100 | 0.18 |
ENSMUST00000111094.8
ENSMUST00000111097.8 |
Fis1
|
fission, mitochondrial 1 |
| chr19_+_56536685 | 0.17 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr7_+_120450406 | 0.17 |
ENSMUST00000143322.9
ENSMUST00000106488.3 |
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr5_-_65650269 | 0.17 |
ENSMUST00000121661.8
|
Smim14
|
small integral membrane protein 14 |
| chr5_-_120950570 | 0.16 |
ENSMUST00000117193.8
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr1_-_171854818 | 0.16 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr19_+_45433899 | 0.15 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr13_+_93441447 | 0.14 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr7_+_89814713 | 0.13 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_-_56536443 | 0.12 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr9_-_78016302 | 0.12 |
ENSMUST00000001402.14
|
Fbxo9
|
f-box protein 9 |
| chrX_-_165992145 | 0.12 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr8_-_41469332 | 0.12 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr8_-_84976330 | 0.11 |
ENSMUST00000019506.9
|
D8Ertd738e
|
DNA segment, Chr 8, ERATO Doi 738, expressed |
| chr3_+_32490525 | 0.11 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr3_+_67799510 | 0.11 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chrX_+_105626790 | 0.11 |
ENSMUST00000101296.9
ENSMUST00000101297.4 |
Gm5127
|
predicted gene 5127 |
| chr19_-_57185988 | 0.11 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr3_-_116505469 | 0.11 |
ENSMUST00000153108.6
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr10_-_129107354 | 0.11 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr1_-_128030148 | 0.10 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr16_-_50151350 | 0.10 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr15_+_6673167 | 0.10 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr1_+_88128323 | 0.10 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr11_+_31823096 | 0.10 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr7_+_30264835 | 0.10 |
ENSMUST00000043850.14
|
Igflr1
|
IGF-like family receptor 1 |
| chr1_-_23948764 | 0.10 |
ENSMUST00000129254.8
|
Smap1
|
small ArfGAP 1 |
| chr19_-_56536646 | 0.09 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr18_+_42669322 | 0.09 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr13_-_81859056 | 0.09 |
ENSMUST00000161920.2
ENSMUST00000048993.12 |
Polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G |
| chr1_-_128256048 | 0.09 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr1_+_55276336 | 0.08 |
ENSMUST00000061334.10
|
Mars2
|
methionine-tRNA synthetase 2 (mitochondrial) |
| chr5_-_87572060 | 0.08 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr1_-_4430481 | 0.08 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_+_109522781 | 0.08 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr5_+_117378510 | 0.08 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr4_+_109092459 | 0.07 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr13_-_48746836 | 0.07 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr13_-_21330037 | 0.06 |
ENSMUST00000216039.3
|
Olfr1368
|
olfactory receptor 1368 |
| chr7_-_108124272 | 0.06 |
ENSMUST00000216919.2
|
Olfr502
|
olfactory receptor 502 |
| chr19_+_34078333 | 0.06 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr14_-_75991903 | 0.06 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr17_-_78991691 | 0.06 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr2_+_91357100 | 0.06 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr2_+_11176891 | 0.06 |
ENSMUST00000028118.9
|
Prkcq
|
protein kinase C, theta |
| chrX_+_56257374 | 0.05 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr19_+_46678336 | 0.05 |
ENSMUST00000074912.8
|
Borcs7
|
BLOC-1 related complex subunit 7 |
| chr3_-_49711706 | 0.05 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr18_-_20995980 | 0.04 |
ENSMUST00000224530.2
|
Trappc8
|
trafficking protein particle complex 8 |
| chr2_+_85648823 | 0.04 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr4_+_109092610 | 0.04 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr1_-_121260298 | 0.04 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr4_+_5724305 | 0.04 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr2_-_88559941 | 0.04 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr6_-_131655849 | 0.04 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr7_+_45271229 | 0.04 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr16_-_75706161 | 0.04 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr8_-_62576140 | 0.04 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr2_-_111452417 | 0.04 |
ENSMUST00000099612.2
|
Olfr1297
|
olfactory receptor 1297 |
| chr9_-_16289527 | 0.03 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr5_-_86437119 | 0.03 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr3_-_49711765 | 0.03 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr9_-_51240201 | 0.03 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr3_-_144555062 | 0.03 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr1_+_180158035 | 0.03 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr7_+_107529653 | 0.03 |
ENSMUST00000217618.3
|
Olfr473
|
olfactory receptor 473 |
| chr16_+_41353360 | 0.03 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr19_-_57185861 | 0.03 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_-_73869071 | 0.03 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr7_+_108312527 | 0.03 |
ENSMUST00000209620.4
ENSMUST00000074730.4 |
Olfr512
|
olfactory receptor 512 |
| chrX_-_133652140 | 0.03 |
ENSMUST00000052431.12
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr12_+_84116099 | 0.03 |
ENSMUST00000046422.11
ENSMUST00000072505.5 |
Acot5
|
acyl-CoA thioesterase 5 |
| chr6_-_131667026 | 0.03 |
ENSMUST00000080619.3
|
Tas2r114
|
taste receptor, type 2, member 114 |
| chr2_+_83642910 | 0.03 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr2_+_88015223 | 0.03 |
ENSMUST00000079599.4
|
Olfr1168
|
olfactory receptor 1168 |
| chr6_+_132844971 | 0.03 |
ENSMUST00000082014.2
|
Tas2r110
|
taste receptor, type 2, member 110 |
| chr14_+_50630215 | 0.03 |
ENSMUST00000058965.4
|
Olfr736
|
olfactory receptor 736 |
| chr10_+_73782857 | 0.03 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr16_-_58850641 | 0.03 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chrX_-_133652080 | 0.03 |
ENSMUST00000113194.8
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr2_+_87574098 | 0.03 |
ENSMUST00000214723.2
|
Olfr1140
|
olfactory receptor 1140 |
| chrX_+_151909893 | 0.03 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr19_-_57185928 | 0.03 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr16_-_22847808 | 0.03 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chrX_+_94942639 | 0.03 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr6_+_108771840 | 0.03 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr5_-_87288177 | 0.02 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr10_+_97482946 | 0.02 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr2_-_87467879 | 0.02 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr2_-_111927126 | 0.02 |
ENSMUST00000207976.4
|
Olfr1314
|
olfactory receptor 1314 |
| chr14_-_50322136 | 0.02 |
ENSMUST00000072370.3
|
Olfr726
|
olfactory receptor 726 |
| chr2_-_87570322 | 0.02 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr5_-_3697806 | 0.02 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr5_+_88523967 | 0.02 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr16_-_19385631 | 0.02 |
ENSMUST00000090062.2
|
Olfr169
|
olfactory receptor 169 |
| chrX_+_9751861 | 0.02 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr2_-_86216760 | 0.02 |
ENSMUST00000102631.2
|
Olfr1058
|
olfactory receptor 1058 |
| chr12_-_41536430 | 0.02 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr2_-_37537224 | 0.02 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr9_-_38577138 | 0.02 |
ENSMUST00000076542.2
|
Olfr917
|
olfactory receptor 917 |
| chr10_+_129336682 | 0.02 |
ENSMUST00000056002.5
ENSMUST00000218901.2 |
Olfr790
|
olfactory receptor 790 |
| chr2_+_85612365 | 0.02 |
ENSMUST00000215945.2
|
Olfr1015
|
olfactory receptor 1015 |
| chr2_+_65499097 | 0.02 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr2_-_86705146 | 0.02 |
ENSMUST00000130722.3
|
Olfr1096
|
olfactory receptor 1096 |
| chr15_+_91722524 | 0.02 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr2_-_85590718 | 0.02 |
ENSMUST00000099916.2
|
Olfr1012
|
olfactory receptor 1012 |
| chr9_-_39515420 | 0.02 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr2_-_88928307 | 0.02 |
ENSMUST00000099789.5
|
Olfr1220
|
olfactory receptor 1220 |
| chr19_-_12300427 | 0.02 |
ENSMUST00000052558.4
|
Olfr1437
|
olfactory receptor 1437 |
| chr3_-_75177378 | 0.02 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr9_+_38240788 | 0.02 |
ENSMUST00000071449.3
|
Olfr25
|
olfactory receptor 25 |
| chr13_-_32960379 | 0.02 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr2_-_87466089 | 0.02 |
ENSMUST00000090711.3
|
Olfr1132
|
olfactory receptor 1132 |
| chr7_+_107501834 | 0.02 |
ENSMUST00000210420.2
|
Olfr472
|
olfactory receptor 472 |
| chr2_-_76700830 | 0.02 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr12_-_118265103 | 0.02 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr6_+_41095752 | 0.02 |
ENSMUST00000103269.3
|
Trbv12-2
|
T cell receptor beta, variable 12-2 |
| chr7_+_107413787 | 0.02 |
ENSMUST00000084756.2
|
Olfr467
|
olfactory receptor 467 |
| chr16_+_84312257 | 0.02 |
ENSMUST00000210306.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr5_-_120950549 | 0.02 |
ENSMUST00000125547.2
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr9_+_38120508 | 0.02 |
ENSMUST00000212815.2
|
Olfr893
|
olfactory receptor 893 |
| chr5_-_87402659 | 0.02 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr3_-_14843512 | 0.02 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr7_+_107916103 | 0.02 |
ENSMUST00000053179.2
|
Olfr491
|
olfactory receptor 491 |
| chr18_+_52748978 | 0.02 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chr7_-_107724503 | 0.02 |
ENSMUST00000210881.2
|
Olfr484
|
olfactory receptor 484 |
| chr7_+_44033520 | 0.02 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr5_+_18167547 | 0.02 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr10_-_23869379 | 0.01 |
ENSMUST00000078532.3
|
Taar7a
|
trace amine-associated receptor 7A |
| chr6_-_98319684 | 0.01 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr17_-_40630096 | 0.01 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr11_+_67061908 | 0.01 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr17_+_14087827 | 0.01 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr7_-_106473450 | 0.01 |
ENSMUST00000060879.6
|
Olfr705
|
olfactory receptor 705 |
| chr11_-_99996452 | 0.01 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr7_+_102576837 | 0.01 |
ENSMUST00000098214.2
|
Olfr572
|
olfactory receptor 572 |
| chr16_-_78887971 | 0.01 |
ENSMUST00000023566.11
ENSMUST00000060402.6 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr7_+_11910120 | 0.01 |
ENSMUST00000062811.6
|
Vmn1r79
|
vomeronasal 1 receptor 79 |
| chr2_+_111311164 | 0.01 |
ENSMUST00000120021.5
|
Olfr1289
|
olfactory receptor 1289 |
| chrX_-_48886577 | 0.01 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr9_-_39484610 | 0.01 |
ENSMUST00000079178.3
|
Olfr959
|
olfactory receptor 959 |
| chr1_-_28819331 | 0.01 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr17_+_38082519 | 0.01 |
ENSMUST00000119082.2
|
Olfr122
|
olfactory receptor 122 |
| chr12_+_31488208 | 0.01 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr10_+_115405891 | 0.01 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr17_+_31652029 | 0.01 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr14_-_123151162 | 0.01 |
ENSMUST00000160401.8
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr16_+_14407073 | 0.01 |
ENSMUST00000100165.4
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr19_+_13142380 | 0.01 |
ENSMUST00000053772.4
|
Olfr1461
|
olfactory receptor 1461 |
| chr7_+_107702486 | 0.01 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chr3_-_144511566 | 0.01 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr6_+_67993691 | 0.01 |
ENSMUST00000103314.3
|
Igkv1-122
|
immunoglobulin kappa chain variable 1-122 |
| chr11_-_55498559 | 0.01 |
ENSMUST00000108853.8
ENSMUST00000075603.5 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr2_-_63014514 | 0.01 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr17_+_31652073 | 0.01 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr2_+_88389795 | 0.01 |
ENSMUST00000090701.5
|
Olfr1188
|
olfactory receptor 1188 |
| chr4_-_112291169 | 0.01 |
ENSMUST00000058605.3
|
Skint9
|
selection and upkeep of intraepithelial T cells 9 |
| chr16_-_58941421 | 0.01 |
ENSMUST00000213228.2
ENSMUST00000216519.2 |
Olfr194
|
olfactory receptor 194 |
| chr9_-_38930915 | 0.01 |
ENSMUST00000073671.3
|
Olfr146
|
olfactory receptor 146 |
| chr10_-_18890281 | 0.01 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr3_+_113824181 | 0.01 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr17_-_37935524 | 0.01 |
ENSMUST00000072265.3
|
Olfr116
|
olfactory receptor 116 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.6 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.5 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 0.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.2 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.4 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.2 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.1 | 0.2 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |