Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
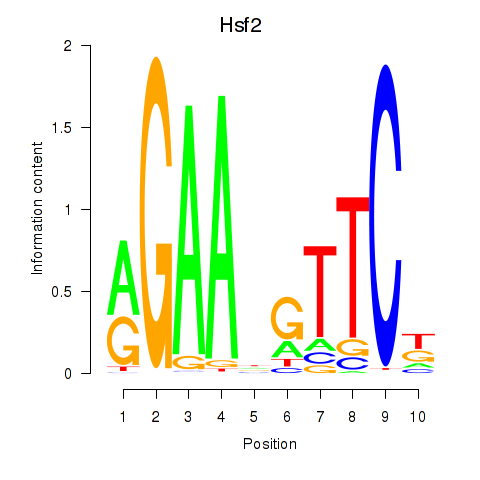
Results for Hsf2
Z-value: 1.12
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSMUSG00000019878.9 | heat shock factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | mm39_v1_chr10_+_57362457_57362510 | 0.99 | 7.6e-04 | Click! |
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_92210348 | 0.75 |
ENSMUST00000032842.13
ENSMUST00000085017.5 |
Ccdc90b
|
coiled-coil domain containing 90B |
| chr18_+_60880149 | 0.69 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr6_+_72575458 | 0.67 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr17_-_34218301 | 0.58 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr4_-_43656437 | 0.56 |
ENSMUST00000030192.5
|
Hint2
|
histidine triad nucleotide binding protein 2 |
| chr14_-_31503869 | 0.49 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr11_-_48836975 | 0.47 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr2_+_29951859 | 0.45 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr7_-_19043955 | 0.42 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chr5_+_104582978 | 0.41 |
ENSMUST00000086833.13
ENSMUST00000031243.15 ENSMUST00000112748.8 ENSMUST00000112746.8 ENSMUST00000145084.8 ENSMUST00000132457.8 |
Spp1
|
secreted phosphoprotein 1 |
| chr11_-_93856783 | 0.41 |
ENSMUST00000021220.10
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr5_+_129864044 | 0.40 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr13_-_111626562 | 0.39 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr13_+_75987987 | 0.39 |
ENSMUST00000022082.8
ENSMUST00000223120.2 ENSMUST00000220523.2 |
Glrx
|
glutaredoxin |
| chr3_-_107925122 | 0.39 |
ENSMUST00000126593.3
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr17_+_35481702 | 0.38 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr7_-_28441173 | 0.38 |
ENSMUST00000171183.2
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr6_-_7692855 | 0.36 |
ENSMUST00000115542.8
ENSMUST00000148349.2 |
Asns
|
asparagine synthetase |
| chr2_+_158148413 | 0.35 |
ENSMUST00000109491.8
ENSMUST00000016168.9 |
Lbp
|
lipopolysaccharide binding protein |
| chr5_+_96941284 | 0.35 |
ENSMUST00000200379.5
|
Anxa3
|
annexin A3 |
| chr1_-_160898181 | 0.35 |
ENSMUST00000035430.4
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr7_-_28441205 | 0.34 |
ENSMUST00000056078.9
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr6_+_51447317 | 0.33 |
ENSMUST00000094623.10
|
Cbx3
|
chromobox 3 |
| chr6_-_129252396 | 0.33 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr17_-_45883421 | 0.33 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr13_+_51799268 | 0.33 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr17_-_26015937 | 0.32 |
ENSMUST00000235925.2
ENSMUST00000238040.2 |
Metrn
|
meteorin, glial cell differentiation regulator |
| chrX_-_72759748 | 0.32 |
ENSMUST00000002091.6
|
Bcap31
|
B cell receptor associated protein 31 |
| chr2_-_30720345 | 0.32 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr2_+_30331839 | 0.32 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr6_-_7693184 | 0.32 |
ENSMUST00000031766.12
|
Asns
|
asparagine synthetase |
| chr12_+_75355082 | 0.31 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr5_+_38233901 | 0.31 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr3_-_115923098 | 0.31 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr17_+_34406523 | 0.31 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr13_+_41154478 | 0.30 |
ENSMUST00000046951.10
|
Pak1ip1
|
PAK1 interacting protein 1 |
| chr12_-_110662677 | 0.29 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_55126882 | 0.29 |
ENSMUST00000021406.6
|
2700097O09Rik
|
RIKEN cDNA 2700097O09 gene |
| chr19_-_4976844 | 0.29 |
ENSMUST00000236496.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr19_+_36325683 | 0.29 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr17_+_34406762 | 0.28 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr1_-_154975376 | 0.28 |
ENSMUST00000055322.6
|
Ier5
|
immediate early response 5 |
| chr10_-_116899664 | 0.28 |
ENSMUST00000218719.2
ENSMUST00000219573.2 ENSMUST00000047672.9 |
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr12_-_110662256 | 0.28 |
ENSMUST00000149189.2
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr14_+_43951187 | 0.28 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr6_-_128415640 | 0.28 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chr15_-_8740218 | 0.28 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_-_26016039 | 0.27 |
ENSMUST00000165838.9
ENSMUST00000002344.7 |
Metrn
|
meteorin, glial cell differentiation regulator |
| chr11_+_115455260 | 0.27 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr2_+_71219561 | 0.27 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr18_+_50186349 | 0.26 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr10_+_130158737 | 0.26 |
ENSMUST00000217702.2
ENSMUST00000042586.10 ENSMUST00000218605.2 |
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr5_+_96941312 | 0.26 |
ENSMUST00000198631.2
|
Anxa3
|
annexin A3 |
| chr4_-_86587728 | 0.26 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr3_+_106020545 | 0.26 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr9_-_14292453 | 0.25 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr5_-_124717055 | 0.25 |
ENSMUST00000128920.2
ENSMUST00000135361.8 |
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chr18_-_43610829 | 0.24 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr19_-_10181243 | 0.24 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr10_+_80662490 | 0.24 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr17_-_36149100 | 0.24 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr19_+_46611826 | 0.23 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr9_-_44318823 | 0.23 |
ENSMUST00000034623.8
|
Trappc4
|
trafficking protein particle complex 4 |
| chr17_-_50401305 | 0.23 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr9_-_44318597 | 0.23 |
ENSMUST00000217163.2
|
Trappc4
|
trafficking protein particle complex 4 |
| chrX_-_135642025 | 0.23 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr1_+_40554513 | 0.23 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chrX_-_135644424 | 0.23 |
ENSMUST00000166478.8
ENSMUST00000113097.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr3_-_107925159 | 0.23 |
ENSMUST00000004140.11
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr9_+_44309727 | 0.23 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr9_+_92132345 | 0.23 |
ENSMUST00000186364.2
|
Plscr1
|
phospholipid scramblase 1 |
| chr2_+_43638814 | 0.23 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr6_+_58810674 | 0.23 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr5_-_5564730 | 0.23 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr10_+_127919142 | 0.23 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr3_+_103078971 | 0.22 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr15_+_100251030 | 0.22 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr7_+_37882642 | 0.22 |
ENSMUST00000178207.10
ENSMUST00000179525.10 |
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr8_+_71047110 | 0.22 |
ENSMUST00000019283.10
ENSMUST00000210005.2 |
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr4_+_88640042 | 0.22 |
ENSMUST00000181601.2
|
Gm26566
|
predicted gene, 26566 |
| chr9_+_5298669 | 0.22 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr13_+_33187205 | 0.21 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr4_+_63478478 | 0.21 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr18_+_46874970 | 0.21 |
ENSMUST00000224622.2
|
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr7_-_141934524 | 0.21 |
ENSMUST00000209263.2
|
Gm49369
|
predicted gene, 49369 |
| chr11_+_69729340 | 0.21 |
ENSMUST00000133967.8
ENSMUST00000094065.5 |
Tmem256
|
transmembrane protein 256 |
| chr17_-_5490504 | 0.20 |
ENSMUST00000005053.14
ENSMUST00000185896.2 ENSMUST00000188282.7 |
Tmem242
|
transmembrane protein 242 |
| chr10_+_7543260 | 0.20 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr15_+_25774070 | 0.20 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr1_+_160898283 | 0.20 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr7_-_30814652 | 0.20 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr18_-_42395207 | 0.20 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr2_+_130119077 | 0.20 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr5_-_23988696 | 0.20 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr6_+_85408953 | 0.20 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr15_+_78810919 | 0.19 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr7_-_24997291 | 0.19 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr5_-_23988551 | 0.19 |
ENSMUST00000148618.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr4_-_120955387 | 0.19 |
ENSMUST00000058754.9
|
Zmpste24
|
zinc metallopeptidase, STE24 |
| chr16_-_33916354 | 0.19 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chrX_-_135641869 | 0.19 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr11_+_53661251 | 0.19 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr2_+_84818538 | 0.19 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chrX_+_7750558 | 0.19 |
ENSMUST00000208640.2
ENSMUST00000207114.2 ENSMUST00000208633.2 ENSMUST00000208397.2 ENSMUST00000153620.3 ENSMUST00000123277.8 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr4_+_133311665 | 0.19 |
ENSMUST00000030661.14
ENSMUST00000105899.2 |
Gpn2
|
GPN-loop GTPase 2 |
| chr1_-_78465479 | 0.19 |
ENSMUST00000190441.2
ENSMUST00000170217.8 ENSMUST00000188247.7 ENSMUST00000068333.14 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr19_+_7534838 | 0.19 |
ENSMUST00000141887.8
ENSMUST00000136756.2 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr2_+_179720416 | 0.19 |
ENSMUST00000087563.7
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr1_+_157334298 | 0.19 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334347 | 0.19 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr8_+_108271643 | 0.19 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_+_44482944 | 0.19 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr5_+_21990251 | 0.18 |
ENSMUST00000239497.2
ENSMUST00000030769.7 |
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chr17_+_13135216 | 0.18 |
ENSMUST00000089024.13
ENSMUST00000151287.8 |
Tcp1
|
t-complex protein 1 |
| chr7_-_126873219 | 0.18 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr5_-_149559636 | 0.18 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_+_93462387 | 0.18 |
ENSMUST00000045756.14
|
S100a10
|
S100 calcium binding protein A10 (calpactin) |
| chr19_-_4889314 | 0.18 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr17_+_35191661 | 0.18 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr15_+_102391614 | 0.18 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chrX_+_7750483 | 0.18 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr16_-_87292592 | 0.18 |
ENSMUST00000176750.2
ENSMUST00000175977.8 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr13_-_19579961 | 0.17 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr5_+_124621227 | 0.17 |
ENSMUST00000136567.2
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr6_-_3988835 | 0.17 |
ENSMUST00000203257.2
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr2_+_29951766 | 0.17 |
ENSMUST00000149578.8
|
Set
|
SET nuclear oncogene |
| chr2_-_154734702 | 0.17 |
ENSMUST00000166171.8
ENSMUST00000161172.3 |
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr10_-_44334711 | 0.17 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr3_+_138566249 | 0.17 |
ENSMUST00000121826.3
|
Tspan5
|
tetraspanin 5 |
| chr7_+_16515265 | 0.17 |
ENSMUST00000108496.9
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr14_-_55897854 | 0.17 |
ENSMUST00000002400.7
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr3_-_106057077 | 0.17 |
ENSMUST00000149836.2
|
Chil3
|
chitinase-like 3 |
| chr1_-_133352115 | 0.17 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr5_+_76331727 | 0.17 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr13_-_19579898 | 0.17 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr19_+_9957940 | 0.17 |
ENSMUST00000235196.2
|
Fth1
|
ferritin heavy polypeptide 1 |
| chr16_-_17711950 | 0.17 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr12_+_111504450 | 0.17 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr6_+_48661503 | 0.16 |
ENSMUST00000067506.14
ENSMUST00000119575.8 ENSMUST00000121957.8 ENSMUST00000090070.6 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr7_+_37883074 | 0.16 |
ENSMUST00000178876.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr10_+_88566918 | 0.16 |
ENSMUST00000116234.9
|
Arl1
|
ADP-ribosylation factor-like 1 |
| chr16_+_4501934 | 0.16 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr12_+_55126999 | 0.16 |
ENSMUST00000220578.2
ENSMUST00000221655.2 |
Srp54a
|
signal recognition particle 54A |
| chr15_-_98729333 | 0.16 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr2_-_24864998 | 0.16 |
ENSMUST00000194392.2
|
Mrpl41
|
mitochondrial ribosomal protein L41 |
| chr7_+_49408847 | 0.16 |
ENSMUST00000085272.7
ENSMUST00000207895.2 |
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr3_-_88821192 | 0.16 |
ENSMUST00000107494.8
|
Msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_-_44140820 | 0.16 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr18_+_47245204 | 0.16 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr1_+_131616407 | 0.16 |
ENSMUST00000064679.9
ENSMUST00000064664.10 ENSMUST00000136247.8 |
Rab7b
|
RAB7B, member RAS oncogene family |
| chr6_+_145067457 | 0.16 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr17_+_47816137 | 0.16 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr9_+_35179153 | 0.16 |
ENSMUST00000034543.5
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr5_-_149559667 | 0.16 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr16_+_19578981 | 0.15 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr13_+_76727787 | 0.15 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr10_-_14581203 | 0.15 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr6_-_120799641 | 0.15 |
ENSMUST00000205049.3
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr5_-_134205559 | 0.15 |
ENSMUST00000076228.3
|
Rcc1l
|
reculator of chromosome condensation 1 like |
| chr11_-_5492175 | 0.15 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr17_+_35780977 | 0.15 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr10_-_130265572 | 0.15 |
ENSMUST00000171811.4
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chr1_+_179788675 | 0.15 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chrX_-_156275231 | 0.15 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr7_+_28050077 | 0.15 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr9_+_106080307 | 0.15 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr6_-_51446752 | 0.15 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr5_+_121342544 | 0.14 |
ENSMUST00000031617.13
|
Rpl6
|
ribosomal protein L6 |
| chr10_+_128073900 | 0.14 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr6_-_136918844 | 0.14 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_-_103609703 | 0.14 |
ENSMUST00000143188.2
|
Caprin1
|
cell cycle associated protein 1 |
| chr18_+_66591604 | 0.14 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr8_+_40876827 | 0.14 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr14_-_66071412 | 0.14 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr19_+_5416769 | 0.14 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr19_+_12438125 | 0.14 |
ENSMUST00000081035.9
|
Mpeg1
|
macrophage expressed gene 1 |
| chr1_+_171173237 | 0.14 |
ENSMUST00000135941.8
|
Pfdn2
|
prefoldin 2 |
| chr16_-_87292711 | 0.14 |
ENSMUST00000176041.8
ENSMUST00000026704.14 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr4_-_116982804 | 0.14 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr3_-_88821297 | 0.14 |
ENSMUST00000126245.2
|
Msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr16_-_18232202 | 0.14 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr17_+_47816074 | 0.14 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr19_-_5416339 | 0.14 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr6_-_30873669 | 0.14 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr4_+_63478454 | 0.14 |
ENSMUST00000124332.8
ENSMUST00000150360.8 |
Tmem268
|
transmembrane protein 268 |
| chr5_-_33940053 | 0.14 |
ENSMUST00000148451.2
ENSMUST00000005431.6 |
Letm1
|
leucine zipper-EF-hand containing transmembrane protein 1 |
| chr6_+_48661477 | 0.14 |
ENSMUST00000118802.8
|
Gimap4
|
GTPase, IMAP family member 4 |
| chr5_+_129924619 | 0.13 |
ENSMUST00000077320.3
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr8_+_106607507 | 0.13 |
ENSMUST00000040254.16
ENSMUST00000119261.8 |
Edc4
|
enhancer of mRNA decapping 4 |
| chr4_+_65523223 | 0.13 |
ENSMUST00000050850.14
ENSMUST00000107366.2 |
Trim32
|
tripartite motif-containing 32 |
| chr7_-_139682295 | 0.13 |
ENSMUST00000120034.8
ENSMUST00000121115.2 ENSMUST00000026539.14 |
Fuom
|
fucose mutarotase |
| chr9_+_75213570 | 0.13 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr19_+_55240357 | 0.13 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr15_+_100659622 | 0.13 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr6_-_129252323 | 0.13 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr8_+_71047010 | 0.13 |
ENSMUST00000211117.2
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr1_-_167112784 | 0.13 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr3_+_32791139 | 0.13 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr7_+_25380583 | 0.13 |
ENSMUST00000108403.4
|
B9d2
|
B9 protein domain 2 |
| chr2_-_181240921 | 0.13 |
ENSMUST00000060173.9
|
Samd10
|
sterile alpha motif domain containing 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 | 0.8 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 0.6 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 0.5 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.3 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.7 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.3 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.3 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.2 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.2 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.2 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.1 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.2 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.2 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.4 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0021679 | cerebellar molecular layer development(GO:0021679) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0033367 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.6 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0045963 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.6 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.1 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0061187 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.0 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 1.0 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.4 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.3 | GO:0016872 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.2 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.1 | 1.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0047726 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |