Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hsf4
Z-value: 2.81
Transcription factors associated with Hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf4
|
ENSMUSG00000033249.11 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf4 | mm39_v1_chr8_+_105996419_105996465 | -0.61 | 2.7e-01 | Click! |
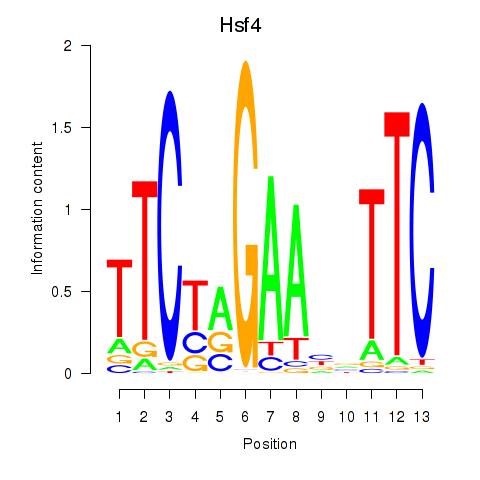
Activity profile of Hsf4 motif
Sorted Z-values of Hsf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_35658131 | 3.12 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr17_+_35643818 | 3.07 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr17_-_34218301 | 2.82 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr12_-_110662256 | 2.57 |
ENSMUST00000149189.2
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr17_+_35643853 | 2.46 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr17_-_45883421 | 2.42 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr15_-_31601652 | 1.97 |
ENSMUST00000161266.2
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr5_+_129864044 | 1.85 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr11_+_70021155 | 1.83 |
ENSMUST00000041550.12
ENSMUST00000165951.8 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chr17_+_35598583 | 1.71 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr6_+_51447317 | 1.69 |
ENSMUST00000094623.10
|
Cbx3
|
chromobox 3 |
| chr17_+_35780977 | 1.68 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr12_+_75355082 | 1.66 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr6_-_128415640 | 1.62 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chr12_-_110662677 | 1.61 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr4_-_136620376 | 1.52 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr6_-_129252396 | 1.49 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr11_-_48836975 | 1.45 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr8_-_85620537 | 1.41 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr17_+_27000034 | 1.38 |
ENSMUST00000015725.16
ENSMUST00000135824.8 ENSMUST00000137989.2 |
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr13_+_28441511 | 1.37 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr12_+_55431007 | 1.37 |
ENSMUST00000163070.8
|
Psma6
|
proteasome subunit alpha 6 |
| chr16_+_22926162 | 1.35 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr5_-_149559636 | 1.26 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr15_-_31601932 | 1.26 |
ENSMUST00000022842.16
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr16_-_84632439 | 1.23 |
ENSMUST00000138279.2
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr5_-_149559667 | 1.23 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_-_45607485 | 1.23 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr15_-_38518406 | 1.23 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr13_-_97897139 | 1.21 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr17_+_35482063 | 1.16 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr11_+_58090382 | 1.13 |
ENSMUST00000035266.11
ENSMUST00000094169.11 ENSMUST00000168280.2 ENSMUST00000058704.9 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr7_+_92210348 | 1.12 |
ENSMUST00000032842.13
ENSMUST00000085017.5 |
Ccdc90b
|
coiled-coil domain containing 90B |
| chr4_-_86587728 | 1.12 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr11_+_22940599 | 1.08 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr5_-_45607463 | 1.08 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr17_+_35481702 | 1.06 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr6_+_88061464 | 1.05 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr5_-_45607554 | 0.99 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr17_+_13135216 | 0.99 |
ENSMUST00000089024.13
ENSMUST00000151287.8 |
Tcp1
|
t-complex protein 1 |
| chr9_-_105398346 | 0.98 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr18_+_66591604 | 0.96 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr7_-_46365108 | 0.93 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr6_+_40302106 | 0.93 |
ENSMUST00000031977.12
|
Agk
|
acylglycerol kinase |
| chr11_+_52123016 | 0.92 |
ENSMUST00000109072.2
|
Skp1
|
S-phase kinase-associated protein 1 |
| chrX_-_72759748 | 0.92 |
ENSMUST00000002091.6
|
Bcap31
|
B cell receptor associated protein 31 |
| chr3_+_159545309 | 0.88 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr11_-_93859064 | 0.86 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr15_-_38518458 | 0.86 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr14_-_36857083 | 0.85 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr6_-_148847854 | 0.84 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr2_-_121287174 | 0.83 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr1_-_33796790 | 0.83 |
ENSMUST00000187602.2
ENSMUST00000044691.9 |
Bag2
|
BCL2-associated athanogene 2 |
| chr16_+_22926504 | 0.80 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr5_+_121342544 | 0.79 |
ENSMUST00000031617.13
|
Rpl6
|
ribosomal protein L6 |
| chr19_-_4675631 | 0.78 |
ENSMUST00000225375.2
ENSMUST00000025823.6 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr10_+_80662490 | 0.78 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr15_-_78377926 | 0.76 |
ENSMUST00000163494.3
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr6_-_129252323 | 0.75 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr2_-_24864998 | 0.75 |
ENSMUST00000194392.2
|
Mrpl41
|
mitochondrial ribosomal protein L41 |
| chr8_+_34621717 | 0.74 |
ENSMUST00000239436.2
ENSMUST00000033933.8 |
Saraf
|
store-operated calcium entry-associated regulatory factor |
| chr11_+_22940519 | 0.73 |
ENSMUST00000173867.8
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr19_-_4675352 | 0.72 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr7_-_120581535 | 0.70 |
ENSMUST00000033169.9
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr8_+_40876827 | 0.69 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr10_+_59159118 | 0.68 |
ENSMUST00000009789.15
ENSMUST00000092512.11 ENSMUST00000105466.3 |
P4ha1
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
| chr16_-_87292592 | 0.68 |
ENSMUST00000176750.2
ENSMUST00000175977.8 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr10_-_79911245 | 0.67 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr11_+_6510167 | 0.67 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr19_-_29790352 | 0.66 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr5_-_130031842 | 0.65 |
ENSMUST00000026613.14
|
Gusb
|
glucuronidase, beta |
| chr10_+_81104006 | 0.65 |
ENSMUST00000057798.9
ENSMUST00000219304.2 |
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr2_+_29678248 | 0.64 |
ENSMUST00000028137.10
ENSMUST00000148791.2 |
Coq4
|
coenzyme Q4 |
| chr13_-_111626562 | 0.64 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr8_-_22888604 | 0.64 |
ENSMUST00000033871.8
|
Slc25a15
|
solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| chr11_+_52122836 | 0.62 |
ENSMUST00000037324.12
ENSMUST00000166537.8 |
Skp1
|
S-phase kinase-associated protein 1 |
| chr5_+_129097133 | 0.62 |
ENSMUST00000031383.14
ENSMUST00000111343.2 |
Ran
|
RAN, member RAS oncogene family |
| chr17_-_13135232 | 0.62 |
ENSMUST00000079121.4
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr2_-_37249247 | 0.62 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr8_-_110419867 | 0.61 |
ENSMUST00000034164.6
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr19_+_5416769 | 0.61 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr8_-_11528615 | 0.60 |
ENSMUST00000033900.7
|
Rab20
|
RAB20, member RAS oncogene family |
| chr2_+_128942919 | 0.60 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr7_-_141472218 | 0.60 |
ENSMUST00000151890.3
|
Tollip
|
toll interacting protein |
| chr9_-_36637670 | 0.60 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr11_-_5492175 | 0.60 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr5_-_88823989 | 0.59 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr11_-_86148379 | 0.59 |
ENSMUST00000132024.8
ENSMUST00000139285.8 |
Ints2
|
integrator complex subunit 2 |
| chr11_-_23447866 | 0.59 |
ENSMUST00000128559.2
ENSMUST00000147157.8 ENSMUST00000109539.8 |
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr19_-_4675300 | 0.59 |
ENSMUST00000225264.2
ENSMUST00000237022.2 ENSMUST00000224675.3 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chrX_+_158086253 | 0.58 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr17_+_25381414 | 0.58 |
ENSMUST00000073277.12
ENSMUST00000182621.8 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr17_-_56428968 | 0.57 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr5_-_23988696 | 0.57 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr6_+_83142902 | 0.56 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr4_-_151142351 | 0.56 |
ENSMUST00000030797.4
|
Vamp3
|
vesicle-associated membrane protein 3 |
| chr9_-_109930078 | 0.56 |
ENSMUST00000196455.2
ENSMUST00000062368.13 |
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr16_-_87292711 | 0.56 |
ENSMUST00000176041.8
ENSMUST00000026704.14 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr6_-_72416531 | 0.55 |
ENSMUST00000205335.2
ENSMUST00000206692.2 ENSMUST00000059472.10 |
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr9_+_71123061 | 0.54 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr11_+_17109263 | 0.54 |
ENSMUST00000102880.5
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
| chr3_-_100876960 | 0.54 |
ENSMUST00000076941.12
|
Ttf2
|
transcription termination factor, RNA polymerase II |
| chr15_+_102391614 | 0.53 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr9_-_36637923 | 0.53 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr2_-_94268426 | 0.53 |
ENSMUST00000028617.7
|
Api5
|
apoptosis inhibitor 5 |
| chr17_+_25992761 | 0.53 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr2_-_51862941 | 0.53 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr8_+_11608412 | 0.52 |
ENSMUST00000209565.2
|
Ing1
|
inhibitor of growth family, member 1 |
| chr19_-_7017295 | 0.52 |
ENSMUST00000025918.9
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr14_+_54457978 | 0.52 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr7_-_19043955 | 0.52 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chr7_+_49408847 | 0.51 |
ENSMUST00000085272.7
ENSMUST00000207895.2 |
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr12_-_110662723 | 0.51 |
ENSMUST00000021698.13
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr7_-_121666486 | 0.51 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr17_-_35265702 | 0.51 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr11_-_23448030 | 0.51 |
ENSMUST00000020529.13
|
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr4_-_86588267 | 0.51 |
ENSMUST00000000466.13
|
Plin2
|
perilipin 2 |
| chr5_-_110220379 | 0.49 |
ENSMUST00000210275.2
|
Gm17655
|
predicted gene, 17655 |
| chr14_-_66071412 | 0.49 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr11_-_121119864 | 0.49 |
ENSMUST00000137299.8
|
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr2_+_158466802 | 0.49 |
ENSMUST00000045644.9
|
Actr5
|
ARP5 actin-related protein 5 |
| chr11_+_5048915 | 0.49 |
ENSMUST00000101610.10
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr17_-_45903494 | 0.49 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr15_-_74671382 | 0.48 |
ENSMUST00000168815.8
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr4_+_65523223 | 0.48 |
ENSMUST00000050850.14
ENSMUST00000107366.2 |
Trim32
|
tripartite motif-containing 32 |
| chr12_-_110662765 | 0.48 |
ENSMUST00000094361.11
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr2_-_29677634 | 0.48 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr16_+_19578981 | 0.47 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr7_-_19005721 | 0.47 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr7_-_89590230 | 0.47 |
ENSMUST00000075010.12
|
Hikeshi
|
heat shock protein nuclear import factor |
| chr4_+_148225139 | 0.47 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr3_+_67337429 | 0.47 |
ENSMUST00000077271.9
|
Gfm1
|
G elongation factor, mitochondrial 1 |
| chr7_-_27713540 | 0.46 |
ENSMUST00000180502.8
|
Zfp850
|
zinc finger protein 850 |
| chr11_-_86148344 | 0.46 |
ENSMUST00000136469.2
ENSMUST00000018212.13 |
Ints2
|
integrator complex subunit 2 |
| chr5_+_149121458 | 0.46 |
ENSMUST00000122160.8
ENSMUST00000100410.10 ENSMUST00000119685.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr5_+_25451771 | 0.46 |
ENSMUST00000144971.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr6_-_51446850 | 0.45 |
ENSMUST00000069949.13
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr2_+_163535925 | 0.45 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr19_-_24938909 | 0.45 |
ENSMUST00000025815.10
|
Cbwd1
|
COBW domain containing 1 |
| chr17_+_21926663 | 0.45 |
ENSMUST00000073312.7
|
Zfp760
|
zinc finger protein 760 |
| chr7_+_136496328 | 0.44 |
ENSMUST00000081510.4
|
Mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr15_+_31602252 | 0.44 |
ENSMUST00000042702.7
ENSMUST00000161061.3 |
Atpsckmt
|
ATP synthase C subunit lysine N-methyltransferase |
| chr5_+_149121355 | 0.43 |
ENSMUST00000050472.16
|
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr9_-_37166699 | 0.43 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr5_-_149559792 | 0.43 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_+_91178288 | 0.43 |
ENSMUST00000171112.8
ENSMUST00000191533.2 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr2_-_35332101 | 0.43 |
ENSMUST00000131745.8
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr10_+_80062468 | 0.42 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr5_-_140375010 | 0.42 |
ENSMUST00000031539.12
|
Snx8
|
sorting nexin 8 |
| chr10_+_28544356 | 0.41 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr10_+_77449543 | 0.41 |
ENSMUST00000124024.3
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr5_-_104059105 | 0.41 |
ENSMUST00000031254.9
|
Klhl8
|
kelch-like 8 |
| chr12_+_83572774 | 0.41 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr1_-_75187417 | 0.41 |
ENSMUST00000113623.8
|
Glb1l
|
galactosidase, beta 1-like |
| chr16_+_33614378 | 0.40 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr2_+_85809620 | 0.39 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr7_+_126575781 | 0.39 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr16_-_84632383 | 0.38 |
ENSMUST00000114191.8
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr12_+_8258166 | 0.38 |
ENSMUST00000220274.2
|
Ldah
|
lipid droplet associated hydrolase |
| chr5_+_149121488 | 0.38 |
ENSMUST00000139474.8
ENSMUST00000117878.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr1_+_59724108 | 0.38 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr6_-_51446752 | 0.37 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr12_-_69274936 | 0.37 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr17_+_31783708 | 0.37 |
ENSMUST00000097352.11
ENSMUST00000237248.2 ENSMUST00000235869.2 ENSMUST00000175806.9 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chr3_+_96537484 | 0.37 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr5_-_129864202 | 0.37 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase |
| chr7_-_47178610 | 0.36 |
ENSMUST00000172559.2
|
Mrgpra2b
|
MAS-related GPR, member A2B |
| chr17_+_29898221 | 0.36 |
ENSMUST00000129864.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr7_+_19248345 | 0.36 |
ENSMUST00000135972.2
|
Trappc6a
|
trafficking protein particle complex 6A |
| chr4_-_133273243 | 0.36 |
ENSMUST00000030665.7
|
Nudc
|
nudC nuclear distribution protein |
| chr11_-_121120052 | 0.35 |
ENSMUST00000169393.8
ENSMUST00000106115.8 ENSMUST00000038709.14 ENSMUST00000147490.6 |
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr9_+_18203421 | 0.35 |
ENSMUST00000001825.9
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr19_+_56414114 | 0.35 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr11_-_70111796 | 0.35 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr4_-_63779562 | 0.35 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr11_+_23206565 | 0.35 |
ENSMUST00000136235.2
|
Xpo1
|
exportin 1 |
| chr10_+_81104041 | 0.35 |
ENSMUST00000218742.2
|
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr9_+_40712562 | 0.34 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr3_+_79498663 | 0.34 |
ENSMUST00000029382.13
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D) |
| chr10_+_77458109 | 0.34 |
ENSMUST00000174510.8
ENSMUST00000172813.2 |
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr1_-_119840887 | 0.34 |
ENSMUST00000037906.6
|
Tmem177
|
transmembrane protein 177 |
| chrX_-_108056995 | 0.34 |
ENSMUST00000033597.9
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr1_-_79836344 | 0.33 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr1_-_55127183 | 0.33 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr8_+_84335176 | 0.33 |
ENSMUST00000212300.2
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr5_+_122988111 | 0.32 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chr1_+_135060994 | 0.32 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr15_-_102097387 | 0.32 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr18_-_70605538 | 0.32 |
ENSMUST00000067556.4
|
4930503L19Rik
|
RIKEN cDNA 4930503L19 gene |
| chr12_-_104964936 | 0.32 |
ENSMUST00000109927.2
ENSMUST00000095439.11 |
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr17_+_25235310 | 0.32 |
ENSMUST00000024983.12
|
Ift140
|
intraflagellar transport 140 |
| chr8_+_27513819 | 0.31 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr6_-_40562700 | 0.31 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chrY_-_90850446 | 0.31 |
ENSMUST00000179623.2
|
Gm21748
|
predicted gene, 21748 |
| chr14_+_66205932 | 0.30 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr9_+_5298669 | 0.30 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr19_+_17114108 | 0.30 |
ENSMUST00000223920.2
ENSMUST00000225351.2 |
Prune2
|
prune homolog 2 |
| chr17_-_74623154 | 0.29 |
ENSMUST00000232687.2
|
Dpy30
|
dpy-30, histone methyltransferase complex regulatory subunit |
| chr1_+_194621127 | 0.29 |
ENSMUST00000016638.8
ENSMUST00000110815.9 |
Cd34
|
CD34 antigen |
| chr2_-_118558825 | 0.29 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.8 | 2.4 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.7 | 5.2 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.6 | 9.2 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.5 | 2.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.5 | 2.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.5 | 1.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.4 | 1.1 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.4 | 1.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 3.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 1.3 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 3.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.3 | 0.9 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 0.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 0.8 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.3 | 1.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.3 | 12.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 0.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.2 | 0.6 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.8 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.2 | 0.5 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 0.5 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 1.5 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 0.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 0.6 | GO:0009838 | abscission(GO:0009838) |
| 0.2 | 0.6 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.1 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.7 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.3 | GO:0097212 | late endosomal microautophagy(GO:0061738) protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.6 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.4 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 1.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.3 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.3 | GO:0072592 | oxygen metabolic process(GO:0072592) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.3 | GO:1901256 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 0.4 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.4 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 1.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.7 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 1.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 1.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.6 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 2.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.5 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.4 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.4 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.7 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0002879 | cell surface pattern recognition receptor signaling pathway(GO:0002752) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 1.0 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.4 | GO:0090077 | foam cell differentiation(GO:0090077) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 17.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.9 | 5.2 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.5 | 9.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.5 | 2.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 0.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 0.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 3.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) germinal vesicle(GO:0042585) |
| 0.0 | 0.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.0 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0080008 | precatalytic spliceosome(GO:0071011) Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 1.1 | 7.6 | GO:0002135 | CTP binding(GO:0002135) |
| 1.1 | 17.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 2.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.4 | 1.3 | GO:0070138 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.4 | 1.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 4.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 1.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 0.9 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 1.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 0.7 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.7 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 0.8 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 0.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 7.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.8 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 1.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.3 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.2 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 1.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 2.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 7.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.5 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 0.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 4.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 2.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 6.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |