Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Ikzf2
Z-value: 0.69
Transcription factors associated with Ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ikzf2
|
ENSMUSG00000025997.14 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf2 | mm39_v1_chr1_-_69724939_69724987 | 0.97 | 7.7e-03 | Click! |
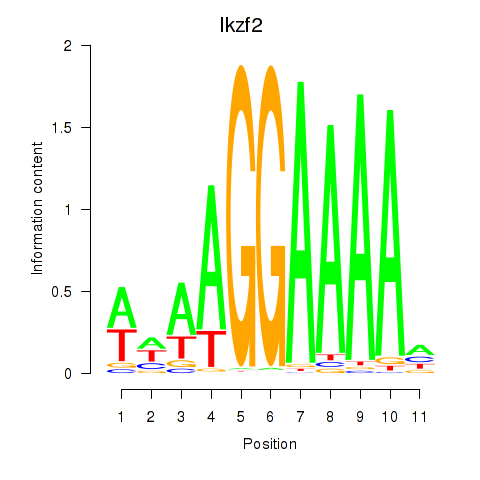
Activity profile of Ikzf2 motif
Sorted Z-values of Ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23759930 | 1.40 |
ENSMUST00000105105.4
|
H3c4
|
H3 clustered histone 4 |
| chr1_-_69724939 | 0.76 |
ENSMUST00000027146.9
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr13_+_23922783 | 0.47 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr4_+_42949814 | 0.46 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr16_-_4031814 | 0.38 |
ENSMUST00000023165.9
|
Crebbp
|
CREB binding protein |
| chr13_+_54738014 | 0.37 |
ENSMUST00000026986.7
|
Higd2a
|
HIG1 domain family, member 2A |
| chr6_-_35110560 | 0.35 |
ENSMUST00000202143.4
ENSMUST00000114993.9 ENSMUST00000114989.9 ENSMUST00000044163.10 ENSMUST00000202417.2 |
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr7_-_126224848 | 0.31 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr1_-_69725045 | 0.31 |
ENSMUST00000190771.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr4_-_44168252 | 0.31 |
ENSMUST00000145760.8
ENSMUST00000128426.8 |
Rnf38
|
ring finger protein 38 |
| chr9_+_123901979 | 0.27 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr15_+_3300249 | 0.26 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr18_-_36859732 | 0.25 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr11_-_96859484 | 0.22 |
ENSMUST00000107623.8
|
Sp2
|
Sp2 transcription factor |
| chr7_+_119495058 | 0.21 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr2_+_174171799 | 0.19 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr10_+_34265969 | 0.18 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr19_-_59931432 | 0.17 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr5_+_42225303 | 0.16 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr16_-_75706161 | 0.16 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr5_+_102629365 | 0.15 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr16_-_95260104 | 0.15 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr9_+_53448322 | 0.15 |
ENSMUST00000035850.8
|
Npat
|
nuclear protein in the AT region |
| chr10_+_107998033 | 0.15 |
ENSMUST00000219263.2
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr9_-_41068771 | 0.14 |
ENSMUST00000136530.8
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr2_-_7400997 | 0.13 |
ENSMUST00000137733.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr6_+_72526236 | 0.13 |
ENSMUST00000114071.8
|
Capg
|
capping protein (actin filament), gelsolin-like |
| chr3_-_95903313 | 0.13 |
ENSMUST00000015889.10
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr2_-_72810782 | 0.13 |
ENSMUST00000102689.10
|
Sp3
|
trans-acting transcription factor 3 |
| chr8_-_81466126 | 0.12 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr3_-_107925159 | 0.11 |
ENSMUST00000004140.11
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr12_+_112645237 | 0.11 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr6_+_115398996 | 0.11 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr5_-_116162415 | 0.11 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr2_-_26299112 | 0.10 |
ENSMUST00000114090.8
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr9_+_123902143 | 0.10 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr5_-_104169696 | 0.10 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr2_-_72810754 | 0.09 |
ENSMUST00000066003.7
|
Sp3
|
trans-acting transcription factor 3 |
| chr3_-_95902949 | 0.09 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr11_+_29413734 | 0.09 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr1_-_39517761 | 0.09 |
ENSMUST00000193823.2
ENSMUST00000054462.11 |
Tbc1d8
|
TBC1 domain family, member 8 |
| chr15_-_83033471 | 0.08 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chrX_+_106193060 | 0.08 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr7_+_119499322 | 0.08 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr6_-_121450547 | 0.08 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr2_-_26299173 | 0.08 |
ENSMUST00000145701.8
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chrX_+_106193167 | 0.08 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr11_+_49500090 | 0.08 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr14_+_53588485 | 0.07 |
ENSMUST00000199280.2
ENSMUST00000103633.2 |
Trav16n
|
T cell receptor alpha variable 16n |
| chrX_+_149981074 | 0.07 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chrX_-_87159237 | 0.07 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr11_-_78074377 | 0.07 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chrX_+_133501928 | 0.06 |
ENSMUST00000074950.11
ENSMUST00000113203.2 ENSMUST00000113202.8 ENSMUST00000050331.13 ENSMUST00000059297.6 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr2_+_112097087 | 0.06 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr5_-_121148143 | 0.06 |
ENSMUST00000202406.4
ENSMUST00000200792.2 |
Rph3a
|
rabphilin 3A |
| chr17_-_45047521 | 0.06 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr5_+_31070739 | 0.06 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr5_+_53747796 | 0.06 |
ENSMUST00000113865.5
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_+_53980561 | 0.06 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr9_-_42855775 | 0.06 |
ENSMUST00000114865.8
|
Grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr3_+_87524848 | 0.05 |
ENSMUST00000039476.15
ENSMUST00000129113.8 |
Arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr5_-_3707166 | 0.05 |
ENSMUST00000196304.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr4_+_42922253 | 0.05 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24 |
| chr6_-_57938488 | 0.05 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr11_-_115382622 | 0.05 |
ENSMUST00000106530.8
ENSMUST00000021082.7 |
Nt5c
|
5',3'-nucleotidase, cytosolic |
| chr1_-_156766351 | 0.05 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_-_121080479 | 0.05 |
ENSMUST00000207968.2
|
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr6_-_128599833 | 0.05 |
ENSMUST00000171306.5
ENSMUST00000204819.3 ENSMUST00000032512.15 |
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr5_-_104169785 | 0.05 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr12_+_98737274 | 0.04 |
ENSMUST00000021399.9
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr17_+_17594808 | 0.04 |
ENSMUST00000232396.2
ENSMUST00000232199.2 |
Riok2
|
RIO kinase 2 |
| chr6_-_128765529 | 0.04 |
ENSMUST00000167691.9
ENSMUST00000174404.8 |
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr12_+_76371634 | 0.04 |
ENSMUST00000154078.3
ENSMUST00000095610.9 |
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr11_+_22970464 | 0.04 |
ENSMUST00000094363.4
ENSMUST00000151877.2 |
Fam161a
|
family with sequence similarity 161, member A |
| chr2_-_77000936 | 0.04 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr12_-_65012270 | 0.04 |
ENSMUST00000222508.2
|
Klhl28
|
kelch-like 28 |
| chr3_-_116762617 | 0.04 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr1_-_87322443 | 0.03 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr2_-_74489763 | 0.03 |
ENSMUST00000173623.2
ENSMUST00000001867.13 |
Evx2
|
even-skipped homeobox 2 |
| chr19_-_13807805 | 0.03 |
ENSMUST00000214475.2
ENSMUST00000067670.5 ENSMUST00000219674.2 ENSMUST00000216287.2 ENSMUST00000215760.2 |
Olfr1500
|
olfactory receptor 1500 |
| chr1_+_66360865 | 0.03 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr15_+_10952418 | 0.03 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr3_+_126157333 | 0.03 |
ENSMUST00000093976.4
|
Arsj
|
arylsulfatase J |
| chr11_-_73290321 | 0.03 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr7_-_139162706 | 0.03 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2 |
| chr19_+_53933271 | 0.03 |
ENSMUST00000025932.9
|
Shoc2
|
Shoc2, leucine rich repeat scaffold protein |
| chr17_+_73144531 | 0.03 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr3_-_116762476 | 0.02 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr8_-_68270870 | 0.02 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr19_-_36097233 | 0.02 |
ENSMUST00000025718.10
|
Ankrd1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr2_-_31006302 | 0.02 |
ENSMUST00000149196.2
|
Fnbp1
|
formin binding protein 1 |
| chr16_-_90524214 | 0.02 |
ENSMUST00000099554.5
|
Mis18a
|
MIS18 kinetochore protein A |
| chr19_-_53932867 | 0.02 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr1_-_162694076 | 0.02 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr6_+_15185202 | 0.02 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr1_-_156766381 | 0.02 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_+_13536696 | 0.02 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr11_-_69692542 | 0.02 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr13_-_21843491 | 0.02 |
ENSMUST00000055615.2
|
Olfr1361
|
olfactory receptor 1361 |
| chr7_-_70010341 | 0.02 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_+_36397426 | 0.02 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr5_-_103359117 | 0.02 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr6_-_128765449 | 0.02 |
ENSMUST00000172601.8
|
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr13_-_57043550 | 0.02 |
ENSMUST00000022023.13
ENSMUST00000174457.9 ENSMUST00000109871.8 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr6_+_55813862 | 0.02 |
ENSMUST00000044729.7
|
Itprid1
|
ITPR interacting domain containing 1 |
| chr5_+_4073343 | 0.02 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr8_-_68658694 | 0.02 |
ENSMUST00000212960.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr19_-_4993060 | 0.02 |
ENSMUST00000133504.2
ENSMUST00000133254.2 ENSMUST00000120475.8 ENSMUST00000025834.15 |
Peli3
|
pellino 3 |
| chr17_-_68311073 | 0.02 |
ENSMUST00000024840.12
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr6_-_85490568 | 0.02 |
ENSMUST00000095759.5
|
Egr4
|
early growth response 4 |
| chr10_+_84591919 | 0.02 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr9_+_59496571 | 0.02 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr17_+_48037758 | 0.02 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr4_+_116983973 | 0.02 |
ENSMUST00000134074.8
|
Dynlt4
|
dynein light chain Tctex-type 4 |
| chr10_-_44334683 | 0.01 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr10_+_25235748 | 0.01 |
ENSMUST00000218903.2
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr12_-_57592907 | 0.01 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr1_+_66361252 | 0.01 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_14374794 | 0.01 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_+_67090878 | 0.01 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr3_+_60436570 | 0.01 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr2_-_77000878 | 0.01 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr1_+_63485002 | 0.01 |
ENSMUST00000087374.10
|
Adam23
|
a disintegrin and metallopeptidase domain 23 |
| chr2_-_71377088 | 0.01 |
ENSMUST00000024159.8
|
Dlx2
|
distal-less homeobox 2 |
| chr3_+_60436163 | 0.01 |
ENSMUST00000194201.6
ENSMUST00000193517.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr19_+_22670134 | 0.01 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_+_105112273 | 0.01 |
ENSMUST00000142230.8
|
Tcl1b2
|
T cell leukemia/lymphoma 1B, 2 |
| chr1_-_57017693 | 0.01 |
ENSMUST00000177282.2
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr6_+_15184630 | 0.01 |
ENSMUST00000115470.3
|
Foxp2
|
forkhead box P2 |
| chr1_-_14374842 | 0.01 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr7_-_89176294 | 0.01 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr5_-_70999547 | 0.01 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr6_-_128599779 | 0.01 |
ENSMUST00000203275.3
|
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr18_+_65715460 | 0.01 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr16_+_41353212 | 0.01 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chrX_-_166907286 | 0.01 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr14_+_53284744 | 0.01 |
ENSMUST00000103606.2
|
Trav16d-dv11
|
T cell receptor alpha variable 16D-DV11 |
| chr2_-_76720097 | 0.01 |
ENSMUST00000148747.8
|
Ttn
|
titin |
| chr7_-_12829100 | 0.01 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr10_+_129308563 | 0.01 |
ENSMUST00000056961.4
|
Olfr788
|
olfactory receptor 788 |
| chr14_+_55173936 | 0.01 |
ENSMUST00000227441.2
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr3_+_159304812 | 0.01 |
ENSMUST00000196999.5
ENSMUST00000029824.9 |
Rpe65
|
retinal pigment epithelium 65 |
| chr6_-_119173699 | 0.01 |
ENSMUST00000239204.2
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chrX_+_6933733 | 0.01 |
ENSMUST00000057101.13
ENSMUST00000115750.8 |
Akap4
|
A kinase (PRKA) anchor protein 4 |
| chrX_+_111003193 | 0.01 |
ENSMUST00000130247.9
ENSMUST00000038546.7 |
Tex16
|
testis expressed gene 16 |
| chr14_+_55173696 | 0.01 |
ENSMUST00000037814.8
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr3_-_69767208 | 0.01 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr8_-_62044164 | 0.01 |
ENSMUST00000135439.2
ENSMUST00000121200.9 |
Palld
|
palladin, cytoskeletal associated protein |
| chr11_-_84416340 | 0.01 |
ENSMUST00000018842.14
|
Lhx1
|
LIM homeobox protein 1 |
| chr2_-_140513382 | 0.01 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chrX_-_4194587 | 0.01 |
ENSMUST00000179325.2
|
Btbd35f20
|
BTB domain containing 35, family member 20 |
| chr12_-_83486669 | 0.01 |
ENSMUST00000177801.2
|
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr12_-_111947536 | 0.00 |
ENSMUST00000185354.2
|
Rd3l
|
retinal degeneration 3-like |
| chr19_+_55882942 | 0.00 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr10_-_129678931 | 0.00 |
ENSMUST00000057775.3
|
Olfr812
|
olfactory receptor 812 |
| chr6_+_92793440 | 0.00 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr2_+_177783713 | 0.00 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr6_+_104470181 | 0.00 |
ENSMUST00000162872.2
|
Cntn6
|
contactin 6 |
| chr7_+_91321500 | 0.00 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr6_+_116533366 | 0.00 |
ENSMUST00000060204.6
|
Olfr214
|
olfactory receptor 214 |
| chr12_+_76371679 | 0.00 |
ENSMUST00000172992.2
|
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr6_+_104469751 | 0.00 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr7_-_133833854 | 0.00 |
ENSMUST00000127524.2
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr16_-_26810402 | 0.00 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr10_+_129298547 | 0.00 |
ENSMUST00000077836.3
|
Olfr787
|
olfactory receptor 787 |
| chr1_+_161322219 | 0.00 |
ENSMUST00000086084.2
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr2_-_86864563 | 0.00 |
ENSMUST00000099868.2
|
Olfr1105
|
olfactory receptor 1105 |
| chr11_-_59181573 | 0.00 |
ENSMUST00000010044.8
|
Wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr8_-_25528972 | 0.00 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr1_-_176041498 | 0.00 |
ENSMUST00000111167.2
|
Pld5
|
phospholipase D family, member 5 |
| chr10_+_101517556 | 0.00 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr17_-_20505815 | 0.00 |
ENSMUST00000167464.3
ENSMUST00000233061.2 ENSMUST00000233933.2 |
Vmn2r106
|
vomeronasal 2, receptor 106 |
| chr6_+_57110440 | 0.00 |
ENSMUST00000226968.2
ENSMUST00000228235.2 |
Vmn1r11
|
vomeronasal 1 receptor 11 |
| chr15_-_96918203 | 0.00 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr11_-_58393380 | 0.00 |
ENSMUST00000170501.3
ENSMUST00000081743.3 |
Olfr331
|
olfactory receptor 331 |
| chr10_-_44334711 | 0.00 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr7_+_140072530 | 0.00 |
ENSMUST00000074897.5
|
Olfr535
|
olfactory receptor 535 |
| chr9_-_13245834 | 0.00 |
ENSMUST00000110582.4
|
Jrkl
|
Jrk-like |
| chr2_-_89193771 | 0.00 |
ENSMUST00000099783.2
|
Olfr1234
|
olfactory receptor 1234 |
| chr10_+_129086405 | 0.00 |
ENSMUST00000097163.3
|
Olfr775
|
olfactory receptor 775 |
| chr6_+_127049865 | 0.00 |
ENSMUST00000000186.9
|
Fgf23
|
fibroblast growth factor 23 |
| chr2_-_140513320 | 0.00 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr7_-_133833734 | 0.00 |
ENSMUST00000134504.8
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr4_-_129472328 | 0.00 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr7_-_106526664 | 0.00 |
ENSMUST00000072368.3
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr5_+_93045837 | 0.00 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr13_+_15637786 | 0.00 |
ENSMUST00000130065.8
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr2_+_87612880 | 0.00 |
ENSMUST00000090709.2
|
Olfr152
|
olfactory receptor 152 |
| chr14_+_53791444 | 0.00 |
ENSMUST00000198297.2
|
Trav14-1
|
T cell receptor alpha variable 14-1 |
| chr2_-_66240408 | 0.00 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr15_-_96917804 | 0.00 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr12_-_111947487 | 0.00 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chr16_-_88571089 | 0.00 |
ENSMUST00000054223.4
|
2310057N15Rik
|
RIKEN cDNA 2310057N15 gene |
| chr11_+_99770013 | 0.00 |
ENSMUST00000078442.4
|
Gm11567
|
predicted gene 11567 |
| chr6_-_23248361 | 0.00 |
ENSMUST00000031709.7
|
Fezf1
|
Fez family zinc finger 1 |
| chr12_+_78243846 | 0.00 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr5_-_51725059 | 0.00 |
ENSMUST00000127135.3
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr11_+_49414548 | 0.00 |
ENSMUST00000077143.7
|
Olfr1383
|
olfactory receptor 1383 |
| chr18_+_13139992 | 0.00 |
ENSMUST00000041676.3
ENSMUST00000234084.2 ENSMUST00000234565.2 |
Hrh4
|
histamine receptor H4 |
| chr4_-_14621497 | 0.00 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0090265 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.1 | 0.4 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 0.2 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |