Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Irf2_Irf1_Irf8_Irf9_Irf7
Z-value: 1.48
Transcription factors associated with Irf2_Irf1_Irf8_Irf9_Irf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
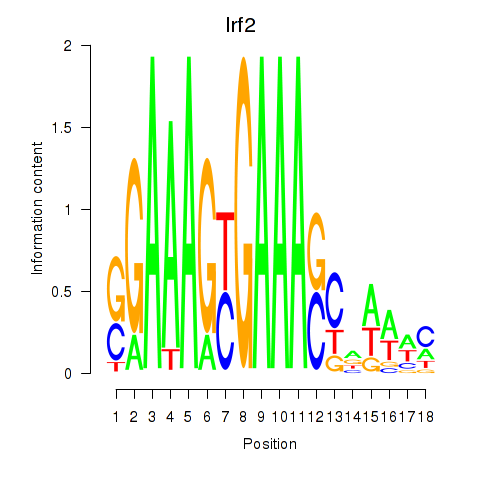
Irf2
|
ENSMUSG00000031627.10 | interferon regulatory factor 2 |
|
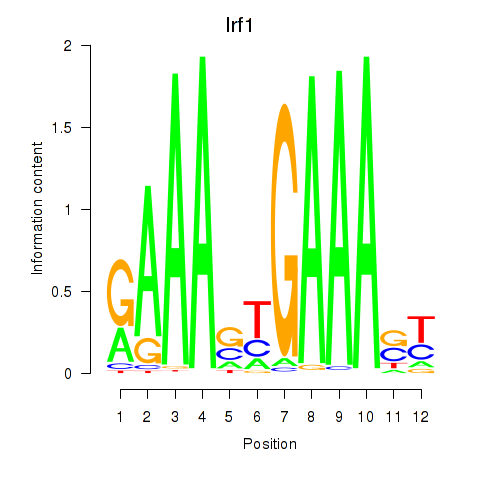
Irf1
|
ENSMUSG00000018899.18 | interferon regulatory factor 1 |
|
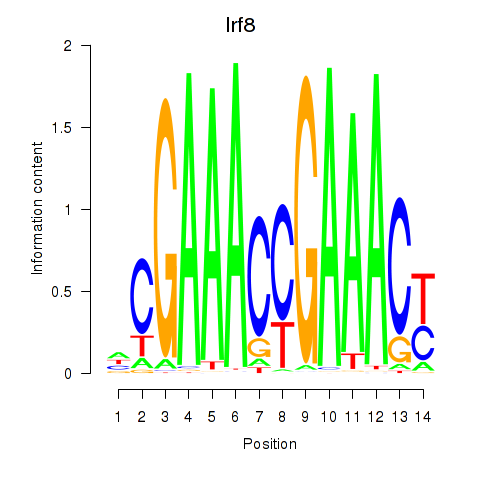
Irf8
|
ENSMUSG00000041515.11 | interferon regulatory factor 8 |
|
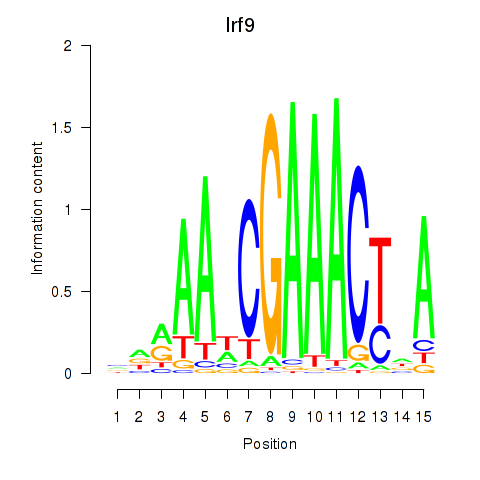
Irf9
|
ENSMUSG00000002325.16 | interferon regulatory factor 9 |
|
Irf7
|
ENSMUSG00000025498.16 | interferon regulatory factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf1 | mm39_v1_chr11_+_53660834_53660886 | 0.98 | 2.7e-03 | Click! |
| Irf2 | mm39_v1_chr8_+_47193275_47193310 | 0.98 | 4.7e-03 | Click! |
| Irf7 | mm39_v1_chr7_-_140845156_140845205 | -0.94 | 1.5e-02 | Click! |
| Irf9 | mm39_v1_chr14_+_55842002_55842036 | 0.93 | 2.2e-02 | Click! |
| Irf8 | mm39_v1_chr8_+_121463090_121463124 | 0.90 | 3.5e-02 | Click! |
Activity profile of Irf2_Irf1_Irf8_Irf9_Irf7 motif
Sorted Z-values of Irf2_Irf1_Irf8_Irf9_Irf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_142326363 | 4.08 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr11_+_48977852 | 2.04 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr12_+_103400470 | 1.68 |
ENSMUST00000079294.12
ENSMUST00000076788.12 ENSMUST00000076702.12 ENSMUST00000066701.13 ENSMUST00000085065.12 ENSMUST00000140838.2 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr17_-_36353582 | 1.42 |
ENSMUST00000058801.15
ENSMUST00000080015.12 ENSMUST00000077960.7 |
H2-T22
|
histocompatibility 2, T region locus 22 |
| chr18_+_60880149 | 1.40 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr11_+_48977888 | 1.35 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr17_-_34406193 | 1.35 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr11_-_48762170 | 1.29 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr8_+_62381115 | 1.27 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr11_-_48883053 | 1.23 |
ENSMUST00000059930.3
ENSMUST00000068063.4 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr2_-_51863203 | 1.17 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr11_-_48955028 | 1.14 |
ENSMUST00000046745.7
|
Tgtp2
|
T cell specific GTPase 2 |
| chr2_-_105229653 | 1.10 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr4_-_40239778 | 1.09 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr15_-_76127600 | 1.03 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr5_-_92496730 | 0.98 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_+_121222865 | 0.88 |
ENSMUST00000032198.11
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr3_+_142265787 | 0.87 |
ENSMUST00000199325.5
ENSMUST00000106222.9 |
Gbp3
|
guanylate binding protein 3 |
| chr6_+_57557978 | 0.87 |
ENSMUST00000031817.10
|
Herc6
|
hect domain and RLD 6 |
| chr6_+_127423779 | 0.86 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr3_+_142265904 | 0.86 |
ENSMUST00000128609.8
ENSMUST00000029935.14 |
Gbp3
|
guanylate binding protein 3 |
| chr3_+_27371168 | 0.85 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr3_+_142266113 | 0.84 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr11_+_119283887 | 0.84 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr16_+_35759346 | 0.83 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr17_+_35658131 | 0.83 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr5_-_105441554 | 0.82 |
ENSMUST00000050011.10
ENSMUST00000196520.2 |
Gm43302
Gbp6
|
predicted gene 43302 guanylate binding protein 6 |
| chr19_+_56385531 | 0.80 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr3_+_142236086 | 0.80 |
ENSMUST00000171263.8
ENSMUST00000045097.11 |
Gbp7
|
guanylate binding protein 7 |
| chr8_-_112064783 | 0.79 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chr8_+_71261073 | 0.78 |
ENSMUST00000000808.8
ENSMUST00000212657.2 ENSMUST00000212146.2 |
Il12rb1
|
interleukin 12 receptor, beta 1 |
| chr9_+_20779924 | 0.77 |
ENSMUST00000043911.8
|
Shfl
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr16_+_23428650 | 0.76 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr9_+_107852733 | 0.75 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr3_+_142202642 | 0.73 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr16_+_35758836 | 0.71 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr13_-_23894697 | 0.69 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr5_-_105287405 | 0.67 |
ENSMUST00000100961.5
ENSMUST00000031235.13 ENSMUST00000197799.2 ENSMUST00000199629.2 ENSMUST00000196677.5 ENSMUST00000100962.8 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr3_+_68598757 | 0.67 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr10_-_34003933 | 0.67 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr5_-_120887864 | 0.66 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr17_+_34406762 | 0.64 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_-_34294377 | 0.64 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr6_+_40619913 | 0.63 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr4_-_40239700 | 0.62 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr6_-_39095144 | 0.62 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr15_-_98626002 | 0.61 |
ENSMUST00000003445.8
|
Fkbp11
|
FK506 binding protein 11 |
| chr11_+_83066009 | 0.60 |
ENSMUST00000000208.13
ENSMUST00000167596.2 |
Slfn4
|
schlafen 4 |
| chr17_+_34406523 | 0.59 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_+_117495337 | 0.59 |
ENSMUST00000031309.16
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chrX_-_94488394 | 0.58 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1 |
| chr2_+_30331839 | 0.58 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr1_-_156501860 | 0.54 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr5_+_115034997 | 0.53 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr17_+_29879569 | 0.53 |
ENSMUST00000024816.13
ENSMUST00000235031.2 ENSMUST00000234911.2 |
Cmtr1
|
cap methyltransferase 1 |
| chr2_-_51862941 | 0.52 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr1_+_153627692 | 0.52 |
ENSMUST00000183241.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr16_-_10603389 | 0.51 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr6_-_125208738 | 0.51 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chr6_+_145067457 | 0.50 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr8_+_95161006 | 0.50 |
ENSMUST00000211816.2
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr8_+_80366247 | 0.49 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chr16_-_22946441 | 0.49 |
ENSMUST00000133847.9
ENSMUST00000115338.8 ENSMUST00000023598.15 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr2_-_25436884 | 0.48 |
ENSMUST00000114234.2
|
Traf2
|
TNF receptor-associated factor 2 |
| chr1_+_52158721 | 0.48 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr13_-_113237505 | 0.48 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr11_+_29668563 | 0.47 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr11_-_100595019 | 0.47 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr9_-_14292453 | 0.47 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr17_-_34218301 | 0.46 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr1_-_37575313 | 0.45 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr7_-_102214636 | 0.45 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr17_-_35081129 | 0.45 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr7_-_144493560 | 0.44 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr7_-_44785603 | 0.44 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr12_-_79054050 | 0.44 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr4_-_156285247 | 0.44 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr11_-_48817986 | 0.44 |
ENSMUST00000094476.9
|
Gm12185
|
predicted gene 12185 |
| chr17_+_29879684 | 0.44 |
ENSMUST00000235014.2
ENSMUST00000130423.4 |
Cmtr1
|
cap methyltransferase 1 |
| chr11_+_72192455 | 0.44 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr8_-_112064393 | 0.44 |
ENSMUST00000145862.3
|
Mlkl
|
mixed lineage kinase domain-like |
| chr13_+_34346936 | 0.44 |
ENSMUST00000124996.8
ENSMUST00000147632.3 |
Psmg4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr17_+_6869070 | 0.43 |
ENSMUST00000092966.5
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr19_-_3955230 | 0.43 |
ENSMUST00000145791.8
|
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr15_+_75734159 | 0.43 |
ENSMUST00000023238.6
ENSMUST00000230514.2 ENSMUST00000229331.2 |
Gsdmd
|
gasdermin D |
| chr3_-_50398027 | 0.43 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr13_-_30170031 | 0.43 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr1_-_173859321 | 0.42 |
ENSMUST00000059226.7
|
Ifi205
|
interferon activated gene 205 |
| chr5_-_105258142 | 0.41 |
ENSMUST00000031238.13
|
Gbp9
|
guanylate-binding protein 9 |
| chr19_+_36387123 | 0.41 |
ENSMUST00000225411.2
ENSMUST00000062389.6 |
Pcgf5
|
polycomb group ring finger 5 |
| chr1_+_52158599 | 0.41 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr19_-_44134872 | 0.40 |
ENSMUST00000237630.2
ENSMUST00000079033.6 |
Bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr17_+_29251602 | 0.40 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr12_+_26519203 | 0.40 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr18_-_43610829 | 0.40 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr12_+_71021395 | 0.40 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr14_-_78970160 | 0.40 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr2_-_132095146 | 0.40 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr8_-_79547707 | 0.40 |
ENSMUST00000130325.8
ENSMUST00000051867.7 |
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_-_80307110 | 0.39 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr2_-_25436952 | 0.39 |
ENSMUST00000028311.13
|
Traf2
|
TNF receptor-associated factor 2 |
| chr9_+_5298669 | 0.38 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr11_-_118292678 | 0.38 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr7_+_138828391 | 0.38 |
ENSMUST00000093993.5
ENSMUST00000172136.9 |
Pwwp2b
|
PWWP domain containing 2B |
| chr17_+_35643818 | 0.38 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr3_+_94350622 | 0.38 |
ENSMUST00000029786.14
ENSMUST00000196143.2 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr17_+_35598583 | 0.37 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr19_+_45549009 | 0.37 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr8_+_79236051 | 0.37 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr1_+_175459559 | 0.36 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr15_+_74593041 | 0.36 |
ENSMUST00000070923.3
|
Them6
|
thioesterase superfamily member 6 |
| chr1_+_52158693 | 0.36 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr3_-_94794256 | 0.36 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr11_+_101339233 | 0.36 |
ENSMUST00000010502.13
|
Ifi35
|
interferon-induced protein 35 |
| chr17_-_34219225 | 0.36 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr11_+_29080733 | 0.36 |
ENSMUST00000020756.9
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr18_+_60345152 | 0.35 |
ENSMUST00000031549.6
|
Gm4951
|
predicted gene 4951 |
| chr11_+_48977495 | 0.35 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr3_-_107667499 | 0.35 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr11_+_58090382 | 0.35 |
ENSMUST00000035266.11
ENSMUST00000094169.11 ENSMUST00000168280.2 ENSMUST00000058704.9 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr10_-_94780695 | 0.34 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr17_+_35643853 | 0.34 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr3_-_104725853 | 0.33 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr17_+_35780977 | 0.33 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr12_-_108859123 | 0.32 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr7_+_103893658 | 0.32 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr5_-_105387395 | 0.32 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr3_-_104725581 | 0.32 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr17_-_35081456 | 0.32 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr2_-_155668567 | 0.31 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr14_-_55828511 | 0.31 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr11_+_69737491 | 0.31 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr16_-_92196954 | 0.31 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr18_+_60509101 | 0.30 |
ENSMUST00000032473.7
ENSMUST00000066912.13 |
Iigp1
|
interferon inducible GTPase 1 |
| chr14_+_55815999 | 0.30 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr13_+_120761861 | 0.30 |
ENSMUST00000225029.2
|
BC147527
|
cDNA sequence BC147527 |
| chr17_-_6923299 | 0.30 |
ENSMUST00000179554.3
|
Dynlt1f
|
dynein light chain Tctex-type 1F |
| chr8_-_106665060 | 0.30 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr18_+_60907698 | 0.29 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr7_-_44888465 | 0.29 |
ENSMUST00000210078.2
|
Cd37
|
CD37 antigen |
| chr2_-_65955338 | 0.29 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr1_-_173770010 | 0.29 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr7_-_112946481 | 0.28 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr7_-_144837719 | 0.28 |
ENSMUST00000058022.6
|
Tpcn2
|
two pore segment channel 2 |
| chr17_+_6697511 | 0.28 |
ENSMUST00000179569.3
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chr18_+_58792514 | 0.28 |
ENSMUST00000025503.10
ENSMUST00000238139.2 |
Isoc1
|
isochorismatase domain containing 1 |
| chr12_-_90705212 | 0.28 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr1_+_85454323 | 0.28 |
ENSMUST00000239236.2
|
Gm7592
|
predicted gene 7592 |
| chr10_-_88192852 | 0.27 |
ENSMUST00000020249.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr19_+_8875459 | 0.27 |
ENSMUST00000096246.5
ENSMUST00000235274.2 |
Ganab
|
alpha glucosidase 2 alpha neutral subunit |
| chr2_+_19376447 | 0.27 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr18_+_60907668 | 0.27 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr19_-_8775817 | 0.27 |
ENSMUST00000235964.2
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr13_+_33188511 | 0.27 |
ENSMUST00000006391.5
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr10_+_39488930 | 0.26 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr4_+_138700195 | 0.26 |
ENSMUST00000123636.8
ENSMUST00000043042.10 ENSMUST00000050949.9 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr2_-_51824656 | 0.26 |
ENSMUST00000165313.2
|
Rbm43
|
RNA binding motif protein 43 |
| chr5_-_120915693 | 0.26 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr1_-_171050004 | 0.26 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr8_+_72943455 | 0.26 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr17_+_37581103 | 0.26 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr11_+_48967411 | 0.26 |
ENSMUST00000109202.3
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr7_+_103893672 | 0.25 |
ENSMUST00000106848.8
|
Trim34a
|
tripartite motif-containing 34A |
| chr3_-_104725535 | 0.25 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr15_-_78947038 | 0.25 |
ENSMUST00000151889.8
ENSMUST00000040676.11 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr16_-_16176390 | 0.25 |
ENSMUST00000115749.3
ENSMUST00000230022.2 |
Dnm1l
|
dynamin 1-like |
| chr7_-_101667346 | 0.25 |
ENSMUST00000209844.2
ENSMUST00000211502.2 ENSMUST00000094134.5 |
Il18bp
|
interleukin 18 binding protein |
| chr5_-_121045568 | 0.24 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr2_+_22785534 | 0.24 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr7_+_80510658 | 0.24 |
ENSMUST00000132163.8
ENSMUST00000205361.2 ENSMUST00000147125.2 |
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr17_+_34138699 | 0.24 |
ENSMUST00000234320.2
|
Tapbp
|
TAP binding protein |
| chr14_+_55815879 | 0.24 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_+_180231038 | 0.24 |
ENSMUST00000029087.4
|
Ogfr
|
opioid growth factor receptor |
| chr7_+_130179063 | 0.24 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr3_+_27371206 | 0.24 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr17_+_35481702 | 0.24 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr1_+_155911451 | 0.23 |
ENSMUST00000111754.9
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr11_-_35725317 | 0.23 |
ENSMUST00000018992.4
|
Rars
|
arginyl-tRNA synthetase |
| chr9_-_14956704 | 0.23 |
ENSMUST00000164273.9
|
Panx1
|
pannexin 1 |
| chr7_-_130148984 | 0.23 |
ENSMUST00000160289.9
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chrX_+_10581248 | 0.23 |
ENSMUST00000144356.8
|
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr19_-_8775935 | 0.22 |
ENSMUST00000096261.5
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr11_-_69572648 | 0.22 |
ENSMUST00000005336.9
|
Senp3
|
SUMO/sentrin specific peptidase 3 |
| chr7_-_84661476 | 0.22 |
ENSMUST00000124773.3
ENSMUST00000233725.2 ENSMUST00000233739.2 ENSMUST00000232837.2 |
Vmn2r66
|
vomeronasal 2, receptor 66 |
| chr13_+_52000704 | 0.21 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr4_+_155666933 | 0.21 |
ENSMUST00000105612.2
|
Nadk
|
NAD kinase |
| chr4_+_106418224 | 0.21 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr7_-_117728790 | 0.21 |
ENSMUST00000206491.2
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr12_-_79027531 | 0.21 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr13_-_23894828 | 0.21 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr7_-_126074222 | 0.21 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr5_-_121025645 | 0.21 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr5_-_134581235 | 0.21 |
ENSMUST00000036999.10
ENSMUST00000100647.7 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr11_+_88890202 | 0.21 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr14_+_55842002 | 0.21 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr5_+_115061293 | 0.21 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr7_-_104157006 | 0.21 |
ENSMUST00000033211.14
ENSMUST00000071069.13 |
Trim30d
|
tripartite motif-containing 30D |
| chr18_-_67378886 | 0.20 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr7_-_126194097 | 0.20 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr16_-_56984137 | 0.20 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf2_Irf1_Irf8_Irf9_Irf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.5 | 1.4 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.4 | 1.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.4 | 1.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.3 | 9.4 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.3 | 1.7 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 2.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 1.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 1.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.3 | 1.1 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 | 0.9 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.4 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.2 | 1.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 0.6 | GO:0090420 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.2 | 2.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.2 | 0.4 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.2 | 0.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.2 | 1.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 7.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.2 | 0.8 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.4 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.1 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:1900063 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.7 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 0.7 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.7 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 2.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.3 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.7 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.1 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.1 | 0.1 | GO:0009138 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.2 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.2 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.6 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.1 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) |
| 0.1 | 0.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.1 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 0.3 | GO:0090669 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 2.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.4 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0019883 | antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.0 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 1.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:1900225 | regulation of NLRP3 inflammasome complex assembly(GO:1900225) |
| 0.0 | 0.0 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 1.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 1.4 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.5 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.1 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 1.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:2000371 | phosphatidylserine biosynthetic process(GO:0006659) regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.2 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:2001245 | negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:1904629 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.0 | 0.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.0 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0006547 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.0 | 0.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.0 | GO:0060809 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.0 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 1.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 0.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.2 | 0.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.2 | 2.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 6.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.3 | 0.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 1.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 4.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 1.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 2.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 0.5 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 2.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.1 | 0.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 1.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.4 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 2.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 2.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.2 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.1 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.1 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 16.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 4.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 1.6 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 8.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |