Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
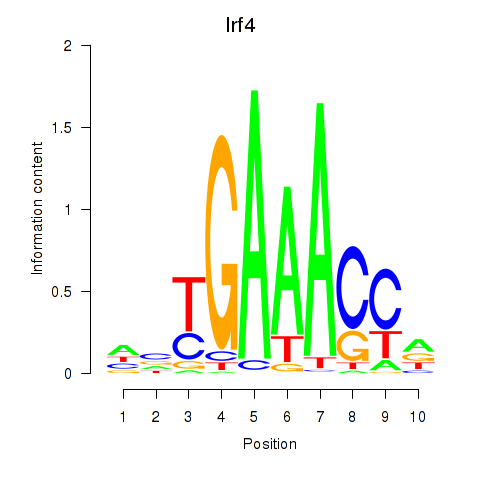
Results for Irf4
Z-value: 1.36
Transcription factors associated with Irf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf4
|
ENSMUSG00000021356.11 | interferon regulatory factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf4 | mm39_v1_chr13_+_30933209_30933290 | -0.64 | 2.4e-01 | Click! |
Activity profile of Irf4 motif
Sorted Z-values of Irf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_142202642 | 1.38 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr11_+_48977852 | 0.97 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr17_+_35780977 | 0.94 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr12_-_110662256 | 0.84 |
ENSMUST00000149189.2
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr10_+_82696135 | 0.83 |
ENSMUST00000219442.3
|
Txnrd1
|
thioredoxin reductase 1 |
| chrX_+_138464065 | 0.78 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr15_-_76127600 | 0.76 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr17_+_35481702 | 0.74 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr6_+_116528102 | 0.74 |
ENSMUST00000122096.3
|
Eif4a3l2
|
eukaryotic translation initiation factor 4A3 like 2 |
| chr11_+_48977888 | 0.71 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chrX_+_93679671 | 0.67 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr2_-_127630769 | 0.63 |
ENSMUST00000028857.14
ENSMUST00000110357.2 |
Nphp1
|
nephronophthisis 1 (juvenile) homolog (human) |
| chr19_-_4976844 | 0.62 |
ENSMUST00000236496.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr19_-_3955230 | 0.62 |
ENSMUST00000145791.8
|
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr17_-_30831576 | 0.58 |
ENSMUST00000235171.2
ENSMUST00000236335.2 ENSMUST00000167624.2 |
Glo1
|
glyoxalase 1 |
| chrM_+_8603 | 0.57 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_+_87938626 | 0.56 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr1_-_160898181 | 0.55 |
ENSMUST00000035430.4
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr3_+_27371168 | 0.55 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr19_+_56385531 | 0.52 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr19_+_46611826 | 0.52 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr13_-_19579898 | 0.50 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr4_-_123033721 | 0.49 |
ENSMUST00000030404.5
|
Ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr18_-_43610829 | 0.47 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_-_136613498 | 0.47 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chrX_+_158086253 | 0.46 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr3_+_89328926 | 0.44 |
ENSMUST00000094378.10
ENSMUST00000137793.2 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr1_+_52158721 | 0.41 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr6_-_39095144 | 0.40 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr1_+_160898283 | 0.40 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr8_+_79236051 | 0.38 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr11_+_87938128 | 0.37 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr17_-_35081456 | 0.37 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr3_+_68598757 | 0.37 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr13_-_19579961 | 0.36 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr11_+_87938519 | 0.36 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr5_-_92496730 | 0.36 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr5_+_24598633 | 0.35 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr4_-_132990362 | 0.35 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr15_+_84921283 | 0.35 |
ENSMUST00000023069.9
|
Fam118a
|
family with sequence similarity 118, member A |
| chr9_+_14187597 | 0.35 |
ENSMUST00000208222.2
|
Sesn3
|
sestrin 3 |
| chr8_+_62381115 | 0.35 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr1_+_52158599 | 0.34 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr19_-_24202344 | 0.34 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr16_+_35759346 | 0.33 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr13_-_23894697 | 0.33 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr15_+_102391614 | 0.32 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr7_-_28441173 | 0.32 |
ENSMUST00000171183.2
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr7_+_79836581 | 0.32 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr10_-_53952686 | 0.31 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr17_-_35081129 | 0.30 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr1_+_52158693 | 0.30 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr11_+_78234300 | 0.29 |
ENSMUST00000002127.14
ENSMUST00000108295.8 |
Unc119
|
unc-119 lipid binding chaperone |
| chr13_-_30170031 | 0.28 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr2_+_29825427 | 0.28 |
ENSMUST00000019859.9
|
Gle1
|
GLE1 RNA export mediator (yeast) |
| chr9_-_35179042 | 0.26 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr7_-_5128936 | 0.26 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr2_+_38401826 | 0.26 |
ENSMUST00000112895.8
|
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr2_-_131016573 | 0.26 |
ENSMUST00000127987.2
|
Spef1
|
sperm flagellar 1 |
| chr8_-_79547707 | 0.25 |
ENSMUST00000130325.8
ENSMUST00000051867.7 |
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr15_-_97603664 | 0.25 |
ENSMUST00000023104.7
|
Rpap3
|
RNA polymerase II associated protein 3 |
| chr3_-_108108113 | 0.25 |
ENSMUST00000106655.2
ENSMUST00000065664.7 |
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr14_+_55815999 | 0.25 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_-_101442663 | 0.23 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr14_+_55815879 | 0.23 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr1_-_97904958 | 0.22 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_-_28441205 | 0.21 |
ENSMUST00000056078.9
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr14_-_78970160 | 0.21 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chrM_+_9459 | 0.21 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr8_-_46577183 | 0.21 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr9_-_107419309 | 0.21 |
ENSMUST00000195235.6
|
Cyb561d2
|
cytochrome b-561 domain containing 2 |
| chr3_+_27371206 | 0.20 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr8_+_105897300 | 0.19 |
ENSMUST00000052209.9
ENSMUST00000109392.9 ENSMUST00000109395.8 |
Cbfb
|
core binding factor beta |
| chr11_-_107361525 | 0.19 |
ENSMUST00000103064.10
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_-_4448631 | 0.19 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr1_+_57445487 | 0.17 |
ENSMUST00000027114.6
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr5_-_92653377 | 0.17 |
ENSMUST00000031377.9
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr14_+_55815580 | 0.17 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr9_-_110886306 | 0.16 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chrX_+_20714782 | 0.16 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chr6_-_49191891 | 0.16 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr18_-_70663209 | 0.16 |
ENSMUST00000161542.8
ENSMUST00000159389.8 |
Poli
|
polymerase (DNA directed), iota |
| chr2_-_152673032 | 0.16 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr6_-_124888643 | 0.16 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr8_-_106665060 | 0.16 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr2_+_22959223 | 0.15 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr3_-_148696155 | 0.15 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_+_109522781 | 0.14 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr2_+_38401655 | 0.14 |
ENSMUST00000054234.10
ENSMUST00000112902.8 |
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr1_-_161704224 | 0.14 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor |
| chr11_+_48977495 | 0.14 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr1_-_75293637 | 0.13 |
ENSMUST00000185419.7
ENSMUST00000187000.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr5_-_104169785 | 0.13 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr2_-_51824656 | 0.13 |
ENSMUST00000165313.2
|
Rbm43
|
RNA binding motif protein 43 |
| chr2_+_22959452 | 0.13 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr13_+_20274708 | 0.12 |
ENSMUST00000072519.7
|
Elmo1
|
engulfment and cell motility 1 |
| chr17_-_7050145 | 0.12 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr6_-_70313491 | 0.12 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr19_-_57185988 | 0.12 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr11_+_78136569 | 0.11 |
ENSMUST00000002133.9
|
Sdf2
|
stromal cell derived factor 2 |
| chr2_+_71283620 | 0.11 |
ENSMUST00000037210.9
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr4_-_4138817 | 0.11 |
ENSMUST00000133567.2
|
Penk
|
preproenkephalin |
| chr2_-_52225146 | 0.11 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr4_-_156285247 | 0.11 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chrX_+_135608725 | 0.11 |
ENSMUST00000055104.6
|
Tceal1
|
transcription elongation factor A (SII)-like 1 |
| chr1_+_74627445 | 0.11 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr13_-_23894828 | 0.10 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr10_+_28544356 | 0.10 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr3_+_152052102 | 0.10 |
ENSMUST00000117492.9
ENSMUST00000026507.13 ENSMUST00000197748.5 |
Usp33
|
ubiquitin specific peptidase 33 |
| chrX_+_149330371 | 0.10 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr7_-_126194097 | 0.10 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr9_+_58536386 | 0.09 |
ENSMUST00000176250.2
|
Nptn
|
neuroplastin |
| chr1_+_175459559 | 0.09 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_+_119283887 | 0.09 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr13_-_100753419 | 0.09 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr14_+_103887644 | 0.08 |
ENSMUST00000069443.14
|
Slain1
|
SLAIN motif family, member 1 |
| chr19_+_41017714 | 0.08 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr10_+_18283405 | 0.07 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr7_+_28441400 | 0.06 |
ENSMUST00000094632.6
|
Sars2
|
seryl-aminoacyl-tRNA synthetase 2 |
| chr1_-_164763091 | 0.06 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr9_+_108385247 | 0.06 |
ENSMUST00000207810.2
ENSMUST00000207862.2 ENSMUST00000208581.2 ENSMUST00000134939.9 ENSMUST00000207713.2 |
Qars
|
glutaminyl-tRNA synthetase |
| chr2_-_66240408 | 0.06 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr1_+_44158111 | 0.06 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr17_-_20334488 | 0.05 |
ENSMUST00000095637.2
|
Fpr-rs7
|
formyl peptide receptor, related sequence 7 |
| chr18_+_37840092 | 0.05 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr7_+_126895531 | 0.05 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr2_+_85597442 | 0.05 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr1_-_75293997 | 0.05 |
ENSMUST00000189282.3
ENSMUST00000191254.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr10_+_21758083 | 0.04 |
ENSMUST00000120509.8
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_+_65504067 | 0.04 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr1_-_75293447 | 0.04 |
ENSMUST00000189551.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr10_+_69048506 | 0.03 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr12_-_114073050 | 0.03 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr7_-_108480811 | 0.03 |
ENSMUST00000059617.2
|
Olfr518
|
olfactory receptor 518 |
| chr1_-_75293942 | 0.03 |
ENSMUST00000187075.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr1_-_57445576 | 0.03 |
ENSMUST00000160837.8
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr19_-_57185861 | 0.03 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr5_-_99391073 | 0.03 |
ENSMUST00000166484.2
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr14_-_61283911 | 0.03 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_-_113047397 | 0.03 |
ENSMUST00000080673.13
ENSMUST00000208151.2 ENSMUST00000208290.2 |
Ryr3
|
ryanodine receptor 3 |
| chr16_-_58930996 | 0.03 |
ENSMUST00000076262.4
|
Olfr193
|
olfactory receptor 193 |
| chr9_-_78350486 | 0.03 |
ENSMUST00000070742.14
ENSMUST00000034898.14 |
Cgas
|
cyclic GMP-AMP synthase |
| chr19_+_47926086 | 0.03 |
ENSMUST00000238163.2
ENSMUST00000066308.9 |
Cfap58
|
cilia and flagella associated protein 58 |
| chr1_-_75293589 | 0.03 |
ENSMUST00000113605.10
|
Dnpep
|
aspartyl aminopeptidase |
| chr9_+_108394761 | 0.03 |
ENSMUST00000192819.2
|
Qrich1
|
glutamine-rich 1 |
| chr7_+_105024231 | 0.02 |
ENSMUST00000106805.4
|
Gm5901
|
predicted gene 5901 |
| chr19_-_57185928 | 0.02 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr11_+_62711057 | 0.02 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr10_+_69048464 | 0.02 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_+_155433858 | 0.02 |
ENSMUST00000080138.13
ENSMUST00000035560.9 ENSMUST00000097529.5 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr11_+_82006001 | 0.02 |
ENSMUST00000009329.3
|
Ccl8
|
chemokine (C-C motif) ligand 8 |
| chr1_-_93029547 | 0.02 |
ENSMUST00000112958.9
ENSMUST00000186861.2 ENSMUST00000171556.8 |
Kif1a
|
kinesin family member 1A |
| chr14_+_65504151 | 0.02 |
ENSMUST00000169656.3
ENSMUST00000226005.2 |
Fbxo16
|
F-box protein 16 |
| chr13_-_100753181 | 0.02 |
ENSMUST00000225754.2
|
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr9_+_5345450 | 0.02 |
ENSMUST00000151332.2
|
Casp12
|
caspase 12 |
| chr10_-_114638202 | 0.02 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr15_-_98707367 | 0.02 |
ENSMUST00000230409.2
|
Ddn
|
dendrin |
| chr11_+_49454750 | 0.02 |
ENSMUST00000204300.2
|
Olfr1380
|
olfactory receptor 1380 |
| chr10_-_111833138 | 0.01 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr2_+_22958956 | 0.01 |
ENSMUST00000226571.2
ENSMUST00000114529.10 ENSMUST00000114526.9 ENSMUST00000228050.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr1_-_75293793 | 0.01 |
ENSMUST00000187836.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr11_+_67212464 | 0.01 |
ENSMUST00000108684.8
|
Myh13
|
myosin, heavy polypeptide 13, skeletal muscle |
| chr19_-_58442866 | 0.01 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_-_57185808 | 0.01 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr10_+_94411119 | 0.01 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr18_+_37637317 | 0.01 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr2_+_111130980 | 0.01 |
ENSMUST00000216697.2
ENSMUST00000213823.2 |
Olfr1279
|
olfactory receptor 1279 |
| chr10_-_52071340 | 0.01 |
ENSMUST00000020045.10
|
Ros1
|
Ros1 proto-oncogene |
| chr19_-_58443012 | 0.01 |
ENSMUST00000129100.8
ENSMUST00000123957.2 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr13_-_113182891 | 0.01 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr3_+_90444537 | 0.01 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr16_-_43484494 | 0.01 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr12_-_118930130 | 0.01 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chrX_-_105884178 | 0.01 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr12_-_113542610 | 0.01 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr11_+_67212604 | 0.01 |
ENSMUST00000180845.8
|
Myh13
|
myosin, heavy polypeptide 13, skeletal muscle |
| chr3_+_90444613 | 0.01 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr11_-_77784922 | 0.01 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr8_+_46338498 | 0.01 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr15_-_101651534 | 0.01 |
ENSMUST00000023710.6
|
Krt71
|
keratin 71 |
| chr7_-_108445215 | 0.01 |
ENSMUST00000071410.2
|
Olfr516
|
olfactory receptor 516 |
| chr18_-_60781365 | 0.01 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin |
| chr9_-_117080869 | 0.01 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr1_+_187995096 | 0.01 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chr17_+_38082519 | 0.01 |
ENSMUST00000119082.2
|
Olfr122
|
olfactory receptor 122 |
| chr19_-_58443830 | 0.01 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr2_-_110591909 | 0.01 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr10_-_107555840 | 0.01 |
ENSMUST00000050702.9
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr15_-_58187556 | 0.01 |
ENSMUST00000022985.2
|
Klhl38
|
kelch-like 38 |
| chr19_+_34618271 | 0.01 |
ENSMUST00000102824.4
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr7_+_58307930 | 0.01 |
ENSMUST00000168747.3
|
Atp10a
|
ATPase, class V, type 10A |
| chr8_+_46338557 | 0.01 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_+_92325386 | 0.00 |
ENSMUST00000029533.3
|
Sprr2j-ps
|
small proline-rich protein 2J, pseudogene |
| chr16_+_65320163 | 0.00 |
ENSMUST00000184525.2
|
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr11_+_53611611 | 0.00 |
ENSMUST00000048605.3
|
Il5
|
interleukin 5 |
| chr10_+_56255184 | 0.00 |
ENSMUST00000220069.2
|
Gja1
|
gap junction protein, alpha 1 |
| chr17_+_23898223 | 0.00 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr1_+_57884693 | 0.00 |
ENSMUST00000169772.3
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr11_+_87457479 | 0.00 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chr2_-_113047313 | 0.00 |
ENSMUST00000208135.2
ENSMUST00000134358.9 |
Ryr3
|
ryanodine receptor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.2 | 0.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.8 | GO:0070212 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.7 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:1904440 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 1.4 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.8 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.8 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.5 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.4 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.6 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.4 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.3 | GO:0097033 | mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.2 | 1.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 2.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.8 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 1.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |