Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Irf5_Irf6
Z-value: 0.36
Transcription factors associated with Irf5_Irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
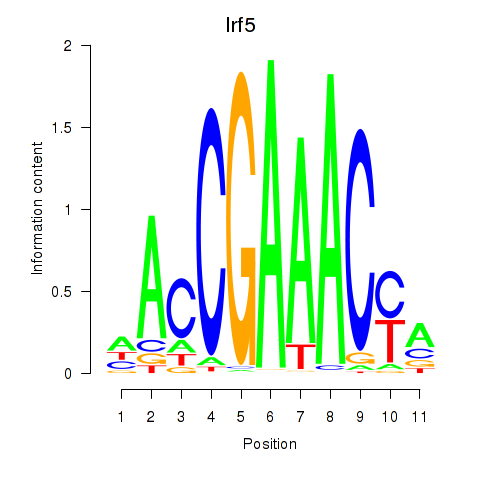
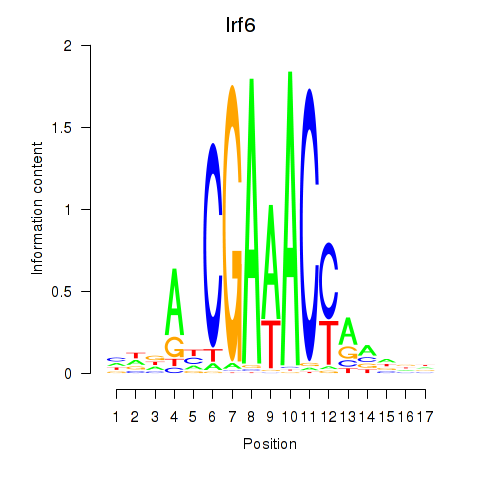
Irf5
|
ENSMUSG00000029771.13 | interferon regulatory factor 5 |
|
Irf6
|
ENSMUSG00000026638.16 | interferon regulatory factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf6 | mm39_v1_chr1_+_192835414_192835516 | 0.78 | 1.2e-01 | Click! |
| Irf5 | mm39_v1_chr6_+_29531247_29531269 | 0.11 | 8.6e-01 | Click! |
Activity profile of Irf5_Irf6 motif
Sorted Z-values of Irf5_Irf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_40239778 | 0.27 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr17_-_79190002 | 0.17 |
ENSMUST00000024884.5
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_+_142202642 | 0.15 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr6_-_124888643 | 0.13 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr8_+_72943455 | 0.13 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr11_+_88890202 | 0.12 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr7_-_125995884 | 0.12 |
ENSMUST00000075671.5
|
Nfatc2ip
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 interacting protein |
| chr6_-_137548004 | 0.11 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr16_-_35691914 | 0.11 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_-_136613498 | 0.11 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr8_+_72050292 | 0.10 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr17_+_35780977 | 0.09 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr14_-_78970160 | 0.09 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr4_-_40239700 | 0.08 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr12_-_26506422 | 0.07 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr3_-_32670628 | 0.07 |
ENSMUST00000193050.2
ENSMUST00000108234.8 ENSMUST00000155737.8 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr9_-_14292453 | 0.07 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr12_-_73160181 | 0.07 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr11_-_86091970 | 0.07 |
ENSMUST00000044423.4
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr16_+_35758836 | 0.06 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr5_+_21391282 | 0.06 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr1_+_175459559 | 0.06 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chrX_+_36390430 | 0.06 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein |
| chr1_+_52158721 | 0.05 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr11_+_101473062 | 0.05 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr11_+_72580823 | 0.05 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr1_+_52158693 | 0.05 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr11_+_72192455 | 0.05 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chrX_-_94488394 | 0.05 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1 |
| chr1_+_52158599 | 0.05 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr16_-_91443794 | 0.05 |
ENSMUST00000232367.2
ENSMUST00000231380.2 ENSMUST00000231444.2 ENSMUST00000232289.2 ENSMUST00000120450.2 ENSMUST00000023684.14 |
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr11_-_94212783 | 0.04 |
ENSMUST00000166312.8
ENSMUST00000107821.9 ENSMUST00000021226.14 ENSMUST00000107820.2 |
Luc7l3
|
LUC7-like 3 (S. cerevisiae) |
| chr17_-_32408431 | 0.04 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chr1_+_175459735 | 0.04 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr7_-_126194097 | 0.04 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr6_+_4505493 | 0.04 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr6_-_39095144 | 0.04 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr5_+_34919078 | 0.04 |
ENSMUST00000080036.3
|
Htt
|
huntingtin |
| chr16_+_23428650 | 0.04 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr19_+_30007910 | 0.03 |
ENSMUST00000025739.14
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chr16_-_35759461 | 0.03 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr9_-_123680726 | 0.03 |
ENSMUST00000084715.14
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr7_-_80037622 | 0.03 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chrX_+_20714782 | 0.02 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chr17_+_34863779 | 0.02 |
ENSMUST00000174796.2
|
Fkbpl
|
FK506 binding protein-like |
| chr7_-_131918926 | 0.02 |
ENSMUST00000080215.6
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr15_-_76127600 | 0.02 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr6_-_70313491 | 0.02 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr5_-_91550853 | 0.02 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr1_-_163141230 | 0.02 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr4_-_118774662 | 0.02 |
ENSMUST00000071979.2
|
Olfr1329
|
olfactory receptor 1329 |
| chr17_-_35081129 | 0.02 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr9_+_44953723 | 0.02 |
ENSMUST00000034600.5
|
Mpzl2
|
myelin protein zero-like 2 |
| chr11_+_60590498 | 0.01 |
ENSMUST00000052346.10
|
Llgl1
|
LLGL1 scribble cell polarity complex component |
| chr11_+_29080733 | 0.01 |
ENSMUST00000020756.9
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr6_+_57557978 | 0.01 |
ENSMUST00000031817.10
|
Herc6
|
hect domain and RLD 6 |
| chr2_-_51824656 | 0.01 |
ENSMUST00000165313.2
|
Rbm43
|
RNA binding motif protein 43 |
| chr4_-_46536088 | 0.01 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr2_-_166904902 | 0.01 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr7_-_65020655 | 0.01 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr12_-_32000534 | 0.01 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr9_+_55234197 | 0.01 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr7_+_127160751 | 0.01 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr1_-_163141278 | 0.01 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr9_+_95836797 | 0.01 |
ENSMUST00000034981.14
ENSMUST00000185633.7 |
Xrn1
|
5'-3' exoribonuclease 1 |
| chr16_+_11131676 | 0.01 |
ENSMUST00000023140.6
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr6_-_146855880 | 0.01 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr13_-_30170031 | 0.01 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr9_-_123680927 | 0.01 |
ENSMUST00000184082.3
ENSMUST00000167595.9 |
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr1_-_105284407 | 0.00 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr16_-_55659194 | 0.00 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr13_-_100689105 | 0.00 |
ENSMUST00000159459.8
|
Ocln
|
occludin |
| chr2_-_77000936 | 0.00 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr17_+_34863738 | 0.00 |
ENSMUST00000036720.9
|
Fkbpl
|
FK506 binding protein-like |
| chr17_+_80597632 | 0.00 |
ENSMUST00000227729.2
ENSMUST00000061703.10 |
Morn2
|
MORN repeat containing 2 |
| chr6_-_13607963 | 0.00 |
ENSMUST00000031554.9
ENSMUST00000149123.3 |
Tmem168
|
transmembrane protein 168 |
| chr6_-_87815653 | 0.00 |
ENSMUST00000204431.2
ENSMUST00000089497.7 |
Isy1
|
ISY1 splicing factor homolog |
| chr9_-_123691077 | 0.00 |
ENSMUST00000182350.3
|
Xcr1
|
chemokine (C motif) receptor 1 |
| chr6_+_70663930 | 0.00 |
ENSMUST00000103402.2
|
Igkv3-3
|
immunoglobulin kappa variable 3-3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf5_Irf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.2 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.3 | GO:1900225 | regulation of NLRP3 inflammasome complex assembly(GO:1900225) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |