Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
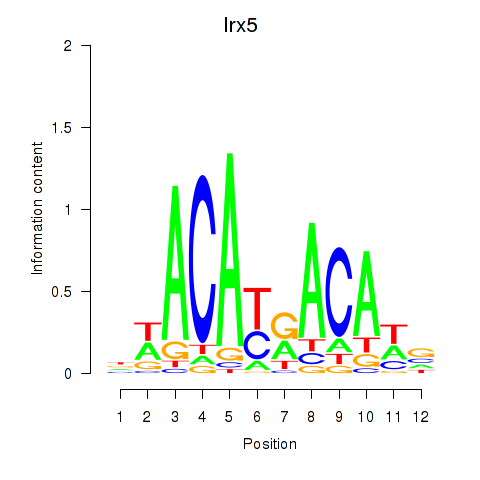
Results for Irx5
Z-value: 0.83
Transcription factors associated with Irx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx5
|
ENSMUSG00000031737.12 | Iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx5 | mm39_v1_chr8_+_93084253_93084379 | -0.79 | 1.1e-01 | Click! |
Activity profile of Irx5 motif
Sorted Z-values of Irx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_68989763 | 0.73 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr12_-_113223839 | 0.52 |
ENSMUST00000194738.6
ENSMUST00000178282.3 |
Igha
|
immunoglobulin heavy constant alpha |
| chr6_+_56974228 | 0.52 |
ENSMUST00000228285.2
ENSMUST00000227847.2 ENSMUST00000226689.2 ENSMUST00000227131.2 ENSMUST00000227631.2 ENSMUST00000227188.2 |
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr4_+_112089442 | 0.50 |
ENSMUST00000038455.12
ENSMUST00000170945.2 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr5_+_107655487 | 0.49 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr11_+_73851643 | 0.48 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr16_+_3408906 | 0.45 |
ENSMUST00000216259.2
|
Olfr161
|
olfactory receptor 161 |
| chr14_+_48358267 | 0.39 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr10_-_129715720 | 0.39 |
ENSMUST00000216966.2
ENSMUST00000213742.2 |
Olfr814
|
olfactory receptor 814 |
| chr8_+_85897951 | 0.37 |
ENSMUST00000076896.6
|
Cks1brt
|
CDC28 protein kinase 1b, retrogene |
| chr11_-_110142565 | 0.36 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr3_+_63203235 | 0.35 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr12_+_71095112 | 0.31 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr6_-_57306479 | 0.30 |
ENSMUST00000227283.2
ENSMUST00000228356.2 |
Vmn1r16
|
vomeronasal 1 receptor 16 |
| chr4_+_148123554 | 0.27 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr7_-_10292412 | 0.27 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr4_-_89229256 | 0.27 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr4_-_131871797 | 0.25 |
ENSMUST00000056336.2
|
Oprd1
|
opioid receptor, delta 1 |
| chr5_-_74189898 | 0.25 |
ENSMUST00000152408.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr19_+_26600820 | 0.24 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_5900130 | 0.24 |
ENSMUST00000026926.5
ENSMUST00000193792.6 ENSMUST00000102981.10 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr10_+_3822667 | 0.23 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr3_+_20011201 | 0.22 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr14_+_50360643 | 0.21 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr4_+_111830119 | 0.21 |
ENSMUST00000106568.8
ENSMUST00000055014.11 ENSMUST00000163281.2 |
Skint7
|
selection and upkeep of intraepithelial T cells 7 |
| chr14_+_54380308 | 0.20 |
ENSMUST00000196323.2
|
Trdc
|
T cell receptor delta, constant region |
| chr18_-_35348049 | 0.19 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr3_-_10273628 | 0.18 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr3_-_126792056 | 0.18 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr12_-_81426238 | 0.15 |
ENSMUST00000062182.8
|
Gm4787
|
predicted gene 4787 |
| chr2_+_172994841 | 0.14 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr9_+_110948492 | 0.12 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_-_87881473 | 0.12 |
ENSMUST00000183862.3
|
Olfr1162
|
olfactory receptor 1162 |
| chr17_-_87332780 | 0.11 |
ENSMUST00000024957.7
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr17_-_35954573 | 0.10 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr7_-_66915756 | 0.10 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr3_+_90173813 | 0.10 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr2_-_89779008 | 0.10 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr19_+_6130059 | 0.09 |
ENSMUST00000149347.8
ENSMUST00000143303.2 |
Tmem262
|
transmembrane protein 262 |
| chr10_+_81128795 | 0.08 |
ENSMUST00000163075.8
ENSMUST00000105327.10 ENSMUST00000045469.15 |
Pip5k1c
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma |
| chr11_-_121410152 | 0.08 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr7_-_86016045 | 0.07 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr6_-_57938488 | 0.07 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr13_+_25127127 | 0.06 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr5_-_46013838 | 0.06 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr11_+_69286473 | 0.06 |
ENSMUST00000144531.2
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr12_-_67269323 | 0.06 |
ENSMUST00000037181.16
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr10_+_86614864 | 0.06 |
ENSMUST00000099396.3
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr11_+_78067537 | 0.05 |
ENSMUST00000098545.12
|
Tlcd1
|
TLC domain containing 1 |
| chr3_-_14843512 | 0.05 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr6_-_113694633 | 0.04 |
ENSMUST00000204533.3
|
Ghrl
|
ghrelin |
| chr13_+_76727799 | 0.04 |
ENSMUST00000109589.3
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr16_-_35951553 | 0.03 |
ENSMUST00000161638.2
ENSMUST00000096090.3 |
Csta1
|
cystatin A1 |
| chr11_+_117006020 | 0.03 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr12_+_85335365 | 0.03 |
ENSMUST00000059341.5
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr8_+_4290105 | 0.02 |
ENSMUST00000110994.9
ENSMUST00000110995.8 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr11_-_69286159 | 0.02 |
ENSMUST00000108660.8
ENSMUST00000051620.5 |
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr3_-_79645101 | 0.02 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr6_+_71470987 | 0.02 |
ENSMUST00000114179.3
|
Rnf103
|
ring finger protein 103 |
| chr1_-_132635042 | 0.02 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr15_-_81742686 | 0.01 |
ENSMUST00000050467.9
ENSMUST00000231000.2 |
Tob2
|
transducer of ERBB2, 2 |
| chr3_+_87265181 | 0.01 |
ENSMUST00000015998.8
|
Cd5l
|
CD5 antigen-like |
| chr16_-_88303859 | 0.01 |
ENSMUST00000069549.3
|
Cldn17
|
claudin 17 |
| chr16_-_44379226 | 0.01 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr6_-_129303659 | 0.01 |
ENSMUST00000203159.2
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr18_-_64794338 | 0.01 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr3_+_138911648 | 0.01 |
ENSMUST00000062306.7
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr14_-_59380335 | 0.00 |
ENSMUST00000022548.10
ENSMUST00000162674.8 ENSMUST00000159858.2 ENSMUST00000162271.2 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr2_+_121991181 | 0.00 |
ENSMUST00000036089.8
|
Trim69
|
tripartite motif-containing 69 |
| chr7_+_55889488 | 0.00 |
ENSMUST00000032633.12
ENSMUST00000155533.2 ENSMUST00000156886.8 |
Oca2
|
oculocutaneous albinism II |
| chr5_-_121641461 | 0.00 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr11_-_99176086 | 0.00 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.3 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |