Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
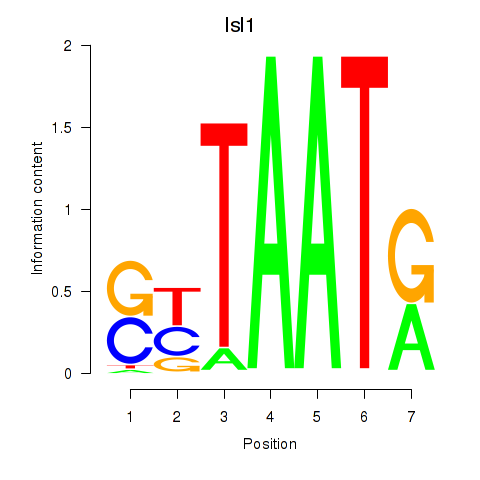
Results for Isl1
Z-value: 1.07
Transcription factors associated with Isl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl1
|
ENSMUSG00000042258.14 | ISL1 transcription factor, LIM/homeodomain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | mm39_v1_chr13_-_116446166_116446235 | -0.63 | 2.6e-01 | Click! |
Activity profile of Isl1 motif
Sorted Z-values of Isl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_43610829 | 0.92 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_-_117715351 | 0.68 |
ENSMUST00000128482.8
ENSMUST00000131840.3 |
Rps15a
|
ribosomal protein S15A |
| chr12_-_73093953 | 0.63 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr11_+_90078485 | 0.53 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr3_+_109481223 | 0.52 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr5_-_77262968 | 0.51 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr9_+_113641615 | 0.45 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr17_-_35265702 | 0.45 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr18_+_35347983 | 0.45 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr13_-_76091931 | 0.43 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr5_-_123663440 | 0.42 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr1_-_10108325 | 0.42 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr15_-_36496880 | 0.40 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr17_+_8384333 | 0.33 |
ENSMUST00000097419.10
ENSMUST00000024636.15 |
Cep43
|
centrosomal protein 43 |
| chr3_-_107952146 | 0.32 |
ENSMUST00000178808.8
ENSMUST00000106670.2 ENSMUST00000029489.15 |
Gstm4
|
glutathione S-transferase, mu 4 |
| chr13_+_76727787 | 0.31 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr7_+_101879176 | 0.30 |
ENSMUST00000120119.9
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr3_+_5815863 | 0.30 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr3_+_134947971 | 0.29 |
ENSMUST00000197369.3
|
Cenpe
|
centromere protein E |
| chr3_+_90383425 | 0.28 |
ENSMUST00000001042.10
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr18_+_77032080 | 0.28 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr14_-_78866714 | 0.27 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr3_-_95662134 | 0.26 |
ENSMUST00000198289.5
ENSMUST00000196868.5 ENSMUST00000074339.13 ENSMUST00000163530.8 ENSMUST00000029752.15 ENSMUST00000195929.5 ENSMUST00000199570.2 ENSMUST00000098857.9 |
Tars2
|
threonyl-tRNA synthetase 2, mitochondrial (putative) |
| chr11_+_78192355 | 0.24 |
ENSMUST00000045026.4
|
Spag5
|
sperm associated antigen 5 |
| chr5_+_44070486 | 0.24 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr1_-_182929025 | 0.24 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr4_-_42168603 | 0.24 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_-_42665763 | 0.23 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr10_+_26648473 | 0.23 |
ENSMUST00000039557.9
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr7_+_140343652 | 0.22 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr14_+_54457978 | 0.21 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr10_+_18345706 | 0.21 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr12_-_25147139 | 0.21 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chrX_+_7446721 | 0.20 |
ENSMUST00000115738.8
|
Foxp3
|
forkhead box P3 |
| chr5_-_43978980 | 0.20 |
ENSMUST00000114047.10
|
Fbxl5
|
F-box and leucine-rich repeat protein 5 |
| chr2_-_71198091 | 0.20 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr10_+_87926932 | 0.20 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr13_+_93444514 | 0.19 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr3_-_37180093 | 0.19 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr10_+_58230203 | 0.19 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_-_86931529 | 0.19 |
ENSMUST00000038422.8
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr10_-_107321938 | 0.17 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr3_+_88461059 | 0.17 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr2_+_120307390 | 0.16 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr18_+_37858753 | 0.16 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr2_+_91376650 | 0.16 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr9_-_44679136 | 0.16 |
ENSMUST00000034607.10
|
Arcn1
|
archain 1 |
| chr3_-_104725853 | 0.15 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr12_-_113802603 | 0.15 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr11_-_76400245 | 0.15 |
ENSMUST00000094012.11
|
Abr
|
active BCR-related gene |
| chr2_+_57887896 | 0.15 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr13_-_103901010 | 0.15 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr1_-_45542442 | 0.14 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr4_+_3940747 | 0.14 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr15_-_34356567 | 0.14 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr2_+_109522781 | 0.14 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr4_-_141412910 | 0.14 |
ENSMUST00000105782.2
|
Rsc1a1
|
regulatory solute carrier protein, family 1, member 1 |
| chr7_+_140796096 | 0.14 |
ENSMUST00000153081.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr17_+_71511642 | 0.13 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr9_+_53678801 | 0.13 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr3_-_148696155 | 0.13 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chrX_+_152020744 | 0.13 |
ENSMUST00000112574.9
|
Klf8
|
Kruppel-like factor 8 |
| chr8_-_41494890 | 0.12 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr1_-_58009191 | 0.12 |
ENSMUST00000159826.2
ENSMUST00000164963.8 |
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr4_+_43406435 | 0.12 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr11_-_4544751 | 0.12 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr8_+_94993453 | 0.12 |
ENSMUST00000212167.2
|
Nup93
|
nucleoporin 93 |
| chr5_+_20112500 | 0.12 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_+_94551929 | 0.12 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr5_+_24679154 | 0.11 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr19_-_40260286 | 0.11 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr14_-_121976273 | 0.10 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr7_+_51537645 | 0.10 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr10_+_28544356 | 0.10 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr4_-_116228921 | 0.10 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr4_+_41966058 | 0.10 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr4_+_150321272 | 0.10 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr9_-_40257586 | 0.10 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr17_-_35454729 | 0.09 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr9_-_82856321 | 0.09 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr4_-_41723129 | 0.09 |
ENSMUST00000171641.2
ENSMUST00000030158.11 |
Dctn3
|
dynactin 3 |
| chr7_-_103094646 | 0.09 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr16_-_21980200 | 0.08 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr10_+_41395870 | 0.08 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr19_+_4036562 | 0.08 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr9_+_35334878 | 0.08 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr2_+_158636727 | 0.08 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr9_-_71803354 | 0.08 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr11_+_4959515 | 0.08 |
ENSMUST00000101613.3
|
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr3_-_49711765 | 0.07 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr5_-_108022900 | 0.07 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr3_-_49711706 | 0.07 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr11_+_117157024 | 0.07 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr6_-_132688891 | 0.07 |
ENSMUST00000105077.2
|
Tas2r122
|
taste receptor, type 2, member 122 |
| chr12_-_113945961 | 0.07 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr2_-_17465410 | 0.07 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr17_-_71153283 | 0.06 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr1_-_58009263 | 0.06 |
ENSMUST00000114410.10
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chrX_+_56257374 | 0.06 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr19_-_37153436 | 0.06 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr3_+_103739877 | 0.06 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr5_+_90666791 | 0.06 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr6_+_125298372 | 0.05 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr4_+_94627513 | 0.05 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr3_+_59914164 | 0.05 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr6_-_54949587 | 0.05 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr11_-_99265721 | 0.05 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr17_-_78991691 | 0.05 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr14_-_50425655 | 0.05 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr11_-_109188892 | 0.05 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr16_+_33650002 | 0.04 |
ENSMUST00000115028.11
ENSMUST00000069345.6 |
Itgb5
|
integrin beta 5 |
| chr1_+_139349912 | 0.04 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr4_-_14621805 | 0.04 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr16_-_88759753 | 0.04 |
ENSMUST00000179707.3
|
Krtap16-3
|
keratin associated protein 16-3 |
| chr1_-_75110511 | 0.04 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr11_-_99412162 | 0.04 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr5_+_20112704 | 0.03 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_-_142716085 | 0.03 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr2_-_88559941 | 0.03 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr3_-_75177378 | 0.03 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chrX_+_162923474 | 0.03 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr18_+_44237577 | 0.03 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr2_+_85600147 | 0.03 |
ENSMUST00000065626.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr1_+_87731360 | 0.03 |
ENSMUST00000177757.2
ENSMUST00000077772.12 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chrX_+_95498965 | 0.03 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr11_-_99412084 | 0.03 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr1_-_170695328 | 0.03 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr19_-_12870888 | 0.03 |
ENSMUST00000216805.3
ENSMUST00000207741.2 |
Olfr1446
|
olfactory receptor 1446 |
| chr2_+_153003212 | 0.03 |
ENSMUST00000089027.3
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr15_-_93417380 | 0.03 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr6_+_132844971 | 0.03 |
ENSMUST00000082014.2
|
Tas2r110
|
taste receptor, type 2, member 110 |
| chr6_-_133014064 | 0.03 |
ENSMUST00000032317.4
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr2_+_29533799 | 0.03 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr19_+_8816663 | 0.03 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr5_+_88523967 | 0.03 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr5_+_149452144 | 0.03 |
ENSMUST00000110502.7
|
Wdr95
|
WD40 repeat domain 95 |
| chr7_+_106413336 | 0.02 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chrX_+_55824797 | 0.02 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr2_-_87805007 | 0.02 |
ENSMUST00000099840.2
|
Olfr74
|
olfactory receptor 74 |
| chr4_-_14621497 | 0.02 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_+_90608751 | 0.02 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chrX_-_42363663 | 0.02 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr10_+_73782857 | 0.02 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr1_+_12788720 | 0.02 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr10_-_18890281 | 0.02 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr6_+_125298296 | 0.02 |
ENSMUST00000081440.14
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr19_-_11816583 | 0.02 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr7_+_143792455 | 0.02 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_+_21506795 | 0.02 |
ENSMUST00000066998.5
|
Vmn1r236
|
vomeronasal 1 receptor 236 |
| chr8_+_55024446 | 0.02 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr2_+_111491764 | 0.02 |
ENSMUST00000213511.2
ENSMUST00000207228.3 ENSMUST00000208983.2 |
Olfr1299
Gm44840
|
olfactory receptor 1299 predicted gene 44840 |
| chr12_-_113790741 | 0.02 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr6_+_142244145 | 0.02 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr3_-_123483772 | 0.02 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr8_-_65489834 | 0.02 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr10_+_28544556 | 0.02 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated |
| chr11_-_99884818 | 0.02 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr1_-_79417732 | 0.02 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr14_+_53775648 | 0.02 |
ENSMUST00000200115.2
ENSMUST00000103650.3 |
Trav12-1
|
T cell receptor alpha variable 12-1 |
| chr2_+_87663385 | 0.02 |
ENSMUST00000077580.2
|
Olfr1148
|
olfactory receptor 1148 |
| chr5_+_81169049 | 0.02 |
ENSMUST00000117253.8
ENSMUST00000120128.8 |
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr3_+_89427458 | 0.02 |
ENSMUST00000000811.8
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr6_-_30936013 | 0.02 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr7_+_119289249 | 0.01 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr2_-_88818544 | 0.01 |
ENSMUST00000099804.2
|
Olfr1214
|
olfactory receptor 1214 |
| chr2_-_111923044 | 0.01 |
ENSMUST00000099598.2
|
Olfr1314
|
olfactory receptor 1314 |
| chr6_+_90105406 | 0.01 |
ENSMUST00000072859.6
|
Vmn1r51
|
vomeronasal 1 receptor 51 |
| chr13_+_111391544 | 0.01 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr2_-_120946877 | 0.01 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr8_-_71964379 | 0.01 |
ENSMUST00000048452.6
|
Plvap
|
plasmalemma vesicle associated protein |
| chr8_-_123187406 | 0.01 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr14_+_10123804 | 0.01 |
ENSMUST00000022262.6
ENSMUST00000224714.2 |
Fezf2
|
Fez family zinc finger 2 |
| chr11_+_46641330 | 0.01 |
ENSMUST00000109223.8
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chr12_-_113823290 | 0.01 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr2_-_89172742 | 0.01 |
ENSMUST00000216561.2
|
Olfr1233
|
olfactory receptor 1233 |
| chr5_+_20433169 | 0.01 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_+_37278647 | 0.01 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr6_-_129428746 | 0.01 |
ENSMUST00000204012.2
ENSMUST00000037481.10 |
Clec1a
|
C-type lectin domain family 1, member a |
| chr2_-_58050494 | 0.01 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr11_-_99313078 | 0.01 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr14_-_118289557 | 0.01 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr2_+_67004178 | 0.01 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr15_+_91722458 | 0.01 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr6_-_144155167 | 0.01 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr18_+_37143758 | 0.01 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chrX_-_151820545 | 0.01 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr10_+_101517556 | 0.01 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr1_-_28819331 | 0.01 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr8_-_62355690 | 0.01 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr9_+_7464139 | 0.01 |
ENSMUST00000217651.3
|
Mmp1a
|
matrix metallopeptidase 1a (interstitial collagenase) |
| chr3_-_144511566 | 0.01 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr10_+_34359513 | 0.01 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chrX_+_113384008 | 0.01 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr18_-_47466378 | 0.01 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr18_+_44467133 | 0.01 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chrX_-_135680890 | 0.01 |
ENSMUST00000018739.5
|
Glra4
|
glycine receptor, alpha 4 subunit |
| chr3_-_7678796 | 0.01 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr5_-_87634665 | 0.01 |
ENSMUST00000201519.2
|
Gm43638
|
predicted gene 43638 |
| chr9_-_95697441 | 0.01 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr6_+_70332836 | 0.01 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.2 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.2 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.3 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.4 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |