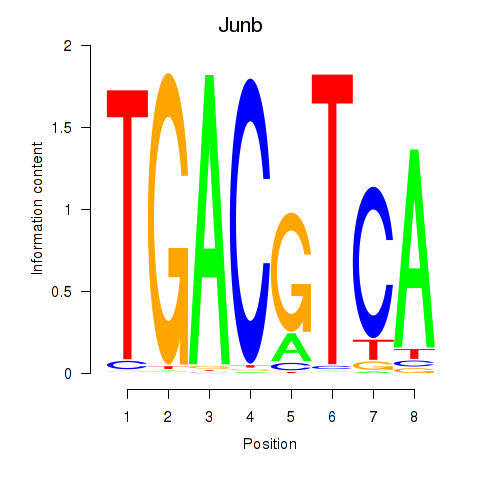
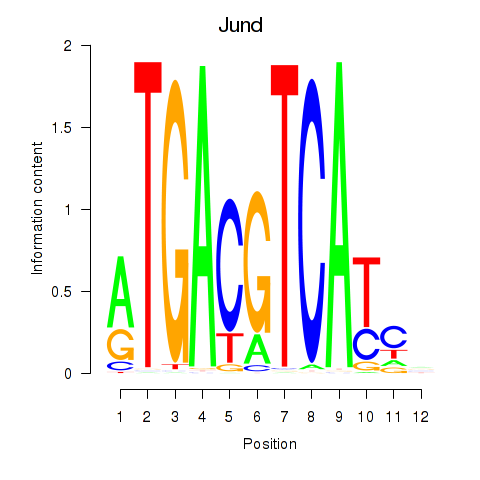
|
chr13_-_54836059
Show fit
|
3.07 |
ENSMUST00000122935.2
ENSMUST00000128257.8
|
Rnf44
|
ring finger protein 44
|
|
chr13_-_54835996
Show fit
|
2.14 |
ENSMUST00000150806.8
ENSMUST00000125927.8
|
Rnf44
|
ring finger protein 44
|
|
chr13_+_23715220
Show fit
|
2.12 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8
|
|
chr13_-_54835878
Show fit
|
1.94 |
ENSMUST00000125871.8
|
Rnf44
|
ring finger protein 44
|
|
chr13_-_54835460
Show fit
|
1.74 |
ENSMUST00000129881.8
|
Rnf44
|
ring finger protein 44
|
|
chr4_-_150736554
Show fit
|
1.69 |
ENSMUST00000117997.2
ENSMUST00000037827.10
|
Slc45a1
|
solute carrier family 45, member 1
|
|
chr1_-_71692320
Show fit
|
1.46 |
ENSMUST00000186940.7
ENSMUST00000188894.7
ENSMUST00000188674.7
ENSMUST00000189821.7
ENSMUST00000187938.7
ENSMUST00000190780.7
ENSMUST00000186736.2
ENSMUST00000055226.13
ENSMUST00000186129.7
|
Fn1
|
fibronectin 1
|
|
chr13_-_54836077
Show fit
|
1.33 |
ENSMUST00000150626.2
ENSMUST00000134177.8
|
Rnf44
|
ring finger protein 44
|
|
chr11_+_87295860
Show fit
|
1.19 |
ENSMUST00000060835.12
|
Tex14
|
testis expressed gene 14
|
|
chr13_-_23945189
Show fit
|
1.19 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1
|
|
chr10_+_121575819
Show fit
|
1.10 |
ENSMUST00000065600.8
ENSMUST00000136432.2
|
BC048403
|
cDNA sequence BC048403
|
|
chr17_+_21503204
Show fit
|
1.05 |
ENSMUST00000236545.2
ENSMUST00000235527.2
|
Vmn1r236
|
vomeronasal 1 receptor 236
|
|
chr7_+_112278520
Show fit
|
1.04 |
ENSMUST00000084705.13
ENSMUST00000239442.2
ENSMUST00000239404.2
ENSMUST00000059768.18
|
Tead1
|
TEA domain family member 1
|
|
chr7_+_24967094
Show fit
|
1.02 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor
|
|
chr19_-_61216834
Show fit
|
1.00 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr11_+_98851238
Show fit
|
0.98 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha
|
|
chr19_+_8828132
Show fit
|
0.97 |
ENSMUST00000235683.2
ENSMUST00000096257.3
|
Lrrn4cl
|
LRRN4 C-terminal like
|
|
chr7_+_16716989
Show fit
|
0.93 |
ENSMUST00000206129.3
|
Gm42372
|
predicted gene, 42372
|
|
chr7_+_44146029
Show fit
|
0.92 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1
|
|
chr17_-_32503689
Show fit
|
0.92 |
ENSMUST00000127893.8
|
Brd4
|
bromodomain containing 4
|
|
chr17_-_32074754
Show fit
|
0.88 |
ENSMUST00000024839.6
|
Sik1
|
salt inducible kinase 1
|
|
chr2_+_30127692
Show fit
|
0.87 |
ENSMUST00000113654.8
ENSMUST00000095078.3
|
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A
|
|
chr19_-_61215743
Show fit
|
0.85 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr7_+_100435548
Show fit
|
0.85 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr13_-_54835508
Show fit
|
0.81 |
ENSMUST00000177950.8
ENSMUST00000146931.8
|
Rnf44
|
ring finger protein 44
|
|
chr12_+_24701273
Show fit
|
0.80 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11
|
|
chr6_+_6863269
Show fit
|
0.78 |
ENSMUST00000171311.8
ENSMUST00000160937.9
|
Dlx6
|
distal-less homeobox 6
|
|
chr16_+_57369595
Show fit
|
0.77 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like
|
|
chr5_-_97259447
Show fit
|
0.76 |
ENSMUST00000069453.9
ENSMUST00000112968.2
|
Paqr3
|
progestin and adipoQ receptor family member III
|
|
chr6_-_149090146
Show fit
|
0.74 |
ENSMUST00000095319.10
ENSMUST00000141346.2
ENSMUST00000111535.8
|
Amn1
|
antagonist of mitotic exit network 1
|
|
chr9_+_45924120
Show fit
|
0.73 |
ENSMUST00000120463.9
ENSMUST00000120247.8
|
Sik3
|
SIK family kinase 3
|
|
chrX_-_20787150
Show fit
|
0.73 |
ENSMUST00000081893.7
ENSMUST00000115345.8
|
Syn1
|
synapsin I
|
|
chr19_-_5135510
Show fit
|
0.72 |
ENSMUST00000140389.8
ENSMUST00000151413.2
ENSMUST00000077066.8
|
Tmem151a
|
transmembrane protein 151A
|
|
chr11_+_68989763
Show fit
|
0.71 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1
|
|
chr8_+_80438421
Show fit
|
0.67 |
ENSMUST00000048147.9
ENSMUST00000210812.2
|
Anapc10
|
anaphase promoting complex subunit 10
|
|
chr9_+_45924105
Show fit
|
0.66 |
ENSMUST00000126865.8
|
Sik3
|
SIK family kinase 3
|
|
chr10_+_53213763
Show fit
|
0.64 |
ENSMUST00000219491.2
ENSMUST00000163319.9
ENSMUST00000220197.2
ENSMUST00000046221.8
ENSMUST00000218468.2
ENSMUST00000219921.2
|
Pln
|
phospholamban
|
|
chr12_+_73333553
Show fit
|
0.64 |
ENSMUST00000140523.8
ENSMUST00000126488.8
|
Slc38a6
|
solute carrier family 38, member 6
|
|
chr4_-_123507494
Show fit
|
0.64 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1
|
|
chr11_+_69656725
Show fit
|
0.63 |
ENSMUST00000108640.8
ENSMUST00000108639.8
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr5_-_124170305
Show fit
|
0.62 |
ENSMUST00000040967.9
|
Vps37b
|
vacuolar protein sorting 37B
|
|
chr13_+_19807274
Show fit
|
0.62 |
ENSMUST00000222464.2
ENSMUST00000002883.7
|
Sfrp4
|
secreted frizzled-related protein 4
|
|
chr4_+_115594951
Show fit
|
0.61 |
ENSMUST00000106522.9
|
Efcab14
|
EF-hand calcium binding domain 14
|
|
chr6_-_39702127
Show fit
|
0.61 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene
|
|
chr8_+_83589979
Show fit
|
0.59 |
ENSMUST00000078525.7
|
Rnf150
|
ring finger protein 150
|
|
chr8_+_66070661
Show fit
|
0.59 |
ENSMUST00000110258.8
ENSMUST00000110256.8
ENSMUST00000110255.8
|
Marchf1
|
membrane associated ring-CH-type finger 1
|
|
chr4_+_32238712
Show fit
|
0.58 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2
|
|
chr16_-_4698148
Show fit
|
0.57 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1
|
|
chr11_+_68986043
Show fit
|
0.56 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1
|
|
chr11_-_58437248
Show fit
|
0.54 |
ENSMUST00000219448.2
|
Olfr329
|
olfactory receptor 329
|
|
chr4_+_85123654
Show fit
|
0.54 |
ENSMUST00000030212.15
ENSMUST00000107189.8
ENSMUST00000107184.8
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr16_+_24540599
Show fit
|
0.53 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_-_103698391
Show fit
|
0.53 |
ENSMUST00000106845.9
ENSMUST00000029438.15
|
Hipk1
|
homeodomain interacting protein kinase 1
|
|
chr13_-_67454476
Show fit
|
0.53 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457
|
|
chr8_+_4375212
Show fit
|
0.53 |
ENSMUST00000127460.8
ENSMUST00000136191.8
|
Ccl25
|
chemokine (C-C motif) ligand 25
|
|
chr13_+_22227359
Show fit
|
0.52 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11
|
|
chr18_-_80756273
Show fit
|
0.52 |
ENSMUST00000078049.12
ENSMUST00000236711.2
|
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1
|
|
chr12_+_73333641
Show fit
|
0.52 |
ENSMUST00000153941.8
ENSMUST00000122920.8
ENSMUST00000101313.4
|
Slc38a6
|
solute carrier family 38, member 6
|
|
chr10_+_29087602
Show fit
|
0.52 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene
|
|
chr6_+_42326980
Show fit
|
0.52 |
ENSMUST00000203849.2
|
Zyx
|
zyxin
|
|
chr7_-_15613769
Show fit
|
0.51 |
ENSMUST00000172758.3
ENSMUST00000044434.13
|
Crx
|
cone-rod homeobox
|
|
chr11_-_70578775
Show fit
|
0.50 |
ENSMUST00000036299.14
ENSMUST00000119120.2
ENSMUST00000100933.10
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr19_+_59249316
Show fit
|
0.49 |
ENSMUST00000026084.5
|
Slc18a2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr9_-_97915036
Show fit
|
0.49 |
ENSMUST00000162295.2
|
Clstn2
|
calsyntenin 2
|
|
chr11_+_69656797
Show fit
|
0.48 |
ENSMUST00000108642.8
ENSMUST00000156932.8
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr4_-_131802606
Show fit
|
0.48 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr1_-_160862364
Show fit
|
0.48 |
ENSMUST00000177003.2
ENSMUST00000159250.9
ENSMUST00000162226.9
|
Zbtb37
|
zinc finger and BTB domain containing 37
|
|
chr7_+_24230063
Show fit
|
0.48 |
ENSMUST00000049020.9
|
Irgq
|
immunity-related GTPase family, Q
|
|
chrX_+_72716756
Show fit
|
0.48 |
ENSMUST00000033752.14
ENSMUST00000114467.9
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr16_+_33504740
Show fit
|
0.48 |
ENSMUST00000232568.2
|
Heg1
|
heart development protein with EGF-like domains 1
|
|
chr6_+_42326760
Show fit
|
0.48 |
ENSMUST00000203652.3
ENSMUST00000070635.13
|
Zyx
|
zyxin
|
|
chr4_+_42949814
Show fit
|
0.47 |
ENSMUST00000037872.10
ENSMUST00000098112.9
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr17_+_35200823
Show fit
|
0.47 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated
|
|
chr8_-_116434517
Show fit
|
0.47 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog
|
|
chr3_+_127347099
Show fit
|
0.47 |
ENSMUST00000043108.9
ENSMUST00000196141.5
ENSMUST00000195955.2
|
Zgrf1
|
zinc finger, GRF-type containing 1
|
|
chr6_+_42326714
Show fit
|
0.46 |
ENSMUST00000203846.3
|
Zyx
|
zyxin
|
|
chr17_-_56783462
Show fit
|
0.46 |
ENSMUST00000067538.6
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S
|
|
chr15_-_58953838
Show fit
|
0.46 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1
|
|
chr4_+_115594918
Show fit
|
0.45 |
ENSMUST00000106525.9
|
Efcab14
|
EF-hand calcium binding domain 14
|
|
chr17_+_20634408
Show fit
|
0.45 |
ENSMUST00000233980.2
ENSMUST00000233642.2
ENSMUST00000233743.2
ENSMUST00000233318.2
ENSMUST00000233161.2
ENSMUST00000233891.2
ENSMUST00000233600.2
ENSMUST00000233863.2
|
Vmn1r224
|
vomeronasal 1 receptor 224
|
|
chrX_-_20928219
Show fit
|
0.44 |
ENSMUST00000040628.6
ENSMUST00000115333.9
ENSMUST00000115334.8
|
Zfp182
|
zinc finger protein 182
|
|
chr9_+_43655230
Show fit
|
0.42 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1
|
|
chr18_-_5334766
Show fit
|
0.42 |
ENSMUST00000234241.2
|
Zfp438
|
zinc finger protein 438
|
|
chr11_-_70578905
Show fit
|
0.41 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr11_-_70578744
Show fit
|
0.41 |
ENSMUST00000108545.9
ENSMUST00000120261.8
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr11_+_79230618
Show fit
|
0.41 |
ENSMUST00000219057.2
ENSMUST00000108251.9
ENSMUST00000071325.9
|
Nf1
|
neurofibromin 1
|
|
chr2_+_28082943
Show fit
|
0.41 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1
|
|
chr17_-_33979280
Show fit
|
0.41 |
ENSMUST00000173860.8
|
Rab11b
|
RAB11B, member RAS oncogene family
|
|
chr13_+_42205491
Show fit
|
0.40 |
ENSMUST00000060148.6
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr8_-_106427696
Show fit
|
0.40 |
ENSMUST00000042608.8
|
Acd
|
adrenocortical dysplasia
|
|
chr2_+_145627900
Show fit
|
0.40 |
ENSMUST00000110005.8
ENSMUST00000094480.11
|
Rin2
|
Ras and Rab interactor 2
|
|
chr17_-_33979442
Show fit
|
0.39 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family
|
|
chr7_-_30443106
Show fit
|
0.39 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic
|
|
chr16_+_95058417
Show fit
|
0.39 |
ENSMUST00000113861.8
ENSMUST00000113854.8
ENSMUST00000113862.8
ENSMUST00000037154.14
ENSMUST00000113855.8
|
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr13_+_5911481
Show fit
|
0.39 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6
|
|
chr11_+_49447705
Show fit
|
0.39 |
ENSMUST00000215360.2
|
Olfr1380
|
olfactory receptor 1380
|
|
chr2_-_164753480
Show fit
|
0.39 |
ENSMUST00000041361.14
|
Zfp335
|
zinc finger protein 335
|
|
chr18_-_35348049
Show fit
|
0.38 |
ENSMUST00000091636.5
ENSMUST00000236680.2
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr1_-_74002156
Show fit
|
0.38 |
ENSMUST00000191367.2
|
Tns1
|
tensin 1
|
|
chr16_+_24212284
Show fit
|
0.38 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_-_101859308
Show fit
|
0.37 |
ENSMUST00000070165.7
ENSMUST00000211235.2
ENSMUST00000211022.2
|
Nup98
|
nucleoporin 98
|
|
chr7_+_125307060
Show fit
|
0.37 |
ENSMUST00000124223.8
ENSMUST00000069660.13
|
Katnip
|
katanin interacting protein
|
|
chr17_-_56783376
Show fit
|
0.36 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S
|
|
chr11_-_6556053
Show fit
|
0.35 |
ENSMUST00000045713.4
|
Nacad
|
NAC alpha domain containing
|
|
chr7_-_141649003
Show fit
|
0.35 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8
|
|
chr2_+_28083105
Show fit
|
0.34 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1
|
|
chrX_+_84091915
Show fit
|
0.34 |
ENSMUST00000239019.2
ENSMUST00000113992.3
ENSMUST00000113991.8
|
Dmd
|
dystrophin, muscular dystrophy
|
|
chr1_-_93029532
Show fit
|
0.34 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A
|
|
chr6_+_83011154
Show fit
|
0.34 |
ENSMUST00000000707.9
ENSMUST00000101257.4
|
Loxl3
|
lysyl oxidase-like 3
|
|
chr17_-_29768586
Show fit
|
0.34 |
ENSMUST00000234305.2
ENSMUST00000234648.2
ENSMUST00000234979.2
|
Gm17657
Tmem217
|
predicted gene, 17657
transmembrane protein 217
|
|
chr4_+_129355374
Show fit
|
0.34 |
ENSMUST00000048162.10
ENSMUST00000138013.3
|
Bsdc1
|
BSD domain containing 1
|
|
chr4_-_89229256
Show fit
|
0.33 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B
|
|
chr7_+_3339077
Show fit
|
0.33 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr5_-_90514061
Show fit
|
0.33 |
ENSMUST00000081914.13
|
Ankrd17
|
ankyrin repeat domain 17
|
|
chr11_+_105866030
Show fit
|
0.33 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr18_+_36481706
Show fit
|
0.33 |
ENSMUST00000235864.2
ENSMUST00000050584.10
|
Cystm1
|
cysteine-rich transmembrane module containing 1
|
|
chr7_+_3339059
Show fit
|
0.32 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr7_+_4140031
Show fit
|
0.32 |
ENSMUST00000128756.8
ENSMUST00000132086.8
ENSMUST00000037472.13
ENSMUST00000117274.8
ENSMUST00000121270.8
ENSMUST00000144248.3
|
Leng8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr17_-_84773544
Show fit
|
0.32 |
ENSMUST00000047524.10
|
Thada
|
thyroid adenoma associated
|
|
chr8_+_3671528
Show fit
|
0.31 |
ENSMUST00000156380.4
|
Pet100
|
PET100 homolog
|
|
chr4_-_44167904
Show fit
|
0.31 |
ENSMUST00000102934.9
|
Rnf38
|
ring finger protein 38
|
|
chr4_-_41275091
Show fit
|
0.31 |
ENSMUST00000030143.13
ENSMUST00000108068.8
|
Ubap2
|
ubiquitin-associated protein 2
|
|
chr18_-_5334349
Show fit
|
0.31 |
ENSMUST00000063989.7
|
Zfp438
|
zinc finger protein 438
|
|
chr12_-_80159768
Show fit
|
0.30 |
ENSMUST00000219642.2
ENSMUST00000165114.2
ENSMUST00000218835.2
ENSMUST00000021552.3
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1
|
|
chr5_+_32293145
Show fit
|
0.30 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2
|
|
chr9_-_20888054
Show fit
|
0.30 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2
|
|
chr11_-_116303791
Show fit
|
0.30 |
ENSMUST00000100202.10
ENSMUST00000106398.9
|
Rnf157
|
ring finger protein 157
|
|
chr10_+_81068980
Show fit
|
0.30 |
ENSMUST00000144087.2
ENSMUST00000117798.8
|
Zfr2
|
zinc finger RNA binding protein 2
|
|
chr16_-_38162174
Show fit
|
0.30 |
ENSMUST00000114740.3
|
Cfap91
|
cilia and flagella associated protein 91
|
|
chr9_+_109760856
Show fit
|
0.30 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4
|
|
chr19_+_5687503
Show fit
|
0.30 |
ENSMUST00000025867.6
|
Rela
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr4_+_107825529
Show fit
|
0.30 |
ENSMUST00000106713.5
ENSMUST00000238795.2
|
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr7_-_142050663
Show fit
|
0.30 |
ENSMUST00000238347.2
|
Prr33
|
proline rich 33
|
|
chr8_-_110464345
Show fit
|
0.29 |
ENSMUST00000212605.2
ENSMUST00000093162.4
ENSMUST00000212726.2
|
Atxn1l
|
ataxin 1-like
|
|
chr9_+_113570580
Show fit
|
0.29 |
ENSMUST00000216817.2
|
Clasp2
|
CLIP associating protein 2
|
|
chr5_-_90514360
Show fit
|
0.29 |
ENSMUST00000168058.7
ENSMUST00000014421.15
|
Ankrd17
|
ankyrin repeat domain 17
|
|
chr3_-_19365431
Show fit
|
0.29 |
ENSMUST00000099195.10
|
Pde7a
|
phosphodiesterase 7A
|
|
chr4_+_8690398
Show fit
|
0.29 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7
|
|
chr13_+_54769611
Show fit
|
0.29 |
ENSMUST00000026991.16
ENSMUST00000137413.8
ENSMUST00000135232.8
ENSMUST00000124752.2
|
Faf2
|
Fas associated factor family member 2
|
|
chr6_+_42326934
Show fit
|
0.29 |
ENSMUST00000203401.3
ENSMUST00000164375.4
|
Zyx
|
zyxin
|
|
chr3_+_144824644
Show fit
|
0.29 |
ENSMUST00000199124.5
|
Odf2l
|
outer dense fiber of sperm tails 2-like
|
|
chrX_+_150303650
Show fit
|
0.29 |
ENSMUST00000112666.8
ENSMUST00000112662.9
ENSMUST00000168501.8
|
Phf8
|
PHD finger protein 8
|
|
chr13_-_64422775
Show fit
|
0.28 |
ENSMUST00000221634.2
ENSMUST00000039318.16
|
Cdc14b
|
CDC14 cell division cycle 14B
|
|
chr9_+_121232480
Show fit
|
0.28 |
ENSMUST00000210351.2
|
Trak1
|
trafficking protein, kinesin binding 1
|
|
chr10_+_59942274
Show fit
|
0.28 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2
|
|
chr16_+_33504908
Show fit
|
0.27 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1
|
|
chr17_+_35201130
Show fit
|
0.27 |
ENSMUST00000173004.2
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated
|
|
chr11_-_70350783
Show fit
|
0.27 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr17_-_45125468
Show fit
|
0.27 |
ENSMUST00000159943.8
ENSMUST00000160673.8
|
Runx2
|
runt related transcription factor 2
|
|
chr17_-_35954573
Show fit
|
0.27 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3
|
|
chr9_+_109760931
Show fit
|
0.27 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4
|
|
chr2_+_90927053
Show fit
|
0.27 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene
|
|
chr2_-_58990967
Show fit
|
0.27 |
ENSMUST00000226455.2
ENSMUST00000077687.6
|
Ccdc148
|
coiled-coil domain containing 148
|
|
chr4_+_56802338
Show fit
|
0.26 |
ENSMUST00000045368.12
|
Abitram
|
actin binding transcription modulator
|
|
chr5_+_104350475
Show fit
|
0.26 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1
|
|
chr10_+_80330669
Show fit
|
0.26 |
ENSMUST00000051773.9
|
Onecut3
|
one cut domain, family member 3
|
|
chr5_-_90514034
Show fit
|
0.26 |
ENSMUST00000218526.2
|
Ankrd17
|
ankyrin repeat domain 17
|
|
chr5_+_123532819
Show fit
|
0.26 |
ENSMUST00000111596.8
ENSMUST00000068237.12
|
Mlxip
|
MLX interacting protein
|
|
chr4_-_131802561
Show fit
|
0.26 |
ENSMUST00000105970.8
ENSMUST00000105975.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr7_+_4140474
Show fit
|
0.26 |
ENSMUST00000154571.7
|
Leng8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr4_-_117354249
Show fit
|
0.26 |
ENSMUST00000030439.15
|
Rnf220
|
ring finger protein 220
|
|
chr1_+_33947250
Show fit
|
0.26 |
ENSMUST00000183034.5
|
Dst
|
dystonin
|
|
chr17_-_43187280
Show fit
|
0.26 |
ENSMUST00000024709.9
ENSMUST00000233476.2
|
Cd2ap
|
CD2-associated protein
|
|
chr17_-_57181420
Show fit
|
0.25 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2
|
|
chr7_-_73187369
Show fit
|
0.25 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2
|
|
chr11_-_101875575
Show fit
|
0.25 |
ENSMUST00000176261.2
ENSMUST00000143177.2
ENSMUST00000003612.13
|
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related)
|
|
chr8_-_112356957
Show fit
|
0.25 |
ENSMUST00000070004.4
|
Ldhd
|
lactate dehydrogenase D
|
|
chr15_-_81284244
Show fit
|
0.25 |
ENSMUST00000172107.8
ENSMUST00000169204.2
ENSMUST00000163382.2
|
St13
|
suppression of tumorigenicity 13
|
|
chr7_-_138447933
Show fit
|
0.25 |
ENSMUST00000118810.2
ENSMUST00000075667.5
ENSMUST00000119664.2
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1
|
|
chr8_-_84996976
Show fit
|
0.25 |
ENSMUST00000005120.12
ENSMUST00000163993.2
ENSMUST00000098578.10
|
Ccdc130
|
coiled-coil domain containing 130
|
|
chr9_+_109760528
Show fit
|
0.25 |
ENSMUST00000035055.15
|
Map4
|
microtubule-associated protein 4
|
|
chr17_-_26309194
Show fit
|
0.25 |
ENSMUST00000237265.2
ENSMUST00000236268.2
ENSMUST00000237875.2
ENSMUST00000150534.9
ENSMUST00000040907.8
|
Decr2
|
2-4-dienoyl-Coenzyme A reductase 2, peroxisomal
|
|
chr7_+_46700349
Show fit
|
0.24 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A
|
|
chr10_+_59942020
Show fit
|
0.24 |
ENSMUST00000121820.9
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2
|
|
chr4_+_134847949
Show fit
|
0.24 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3
|
|
chr5_-_25428151
Show fit
|
0.24 |
ENSMUST00000066954.2
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene
|
|
chr10_-_30076488
Show fit
|
0.24 |
ENSMUST00000216853.2
|
Cenpw
|
centromere protein W
|
|
chr1_-_134006847
Show fit
|
0.24 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2
|
|
chr17_-_36290571
Show fit
|
0.23 |
ENSMUST00000173724.2
ENSMUST00000172900.8
ENSMUST00000174849.8
|
Prr3
|
proline-rich polypeptide 3
|
|
chr11_+_79551358
Show fit
|
0.23 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II)
|
|
chr7_+_125307116
Show fit
|
0.23 |
ENSMUST00000148701.4
|
Katnip
|
katanin interacting protein
|
|
chr13_-_100240570
Show fit
|
0.23 |
ENSMUST00000038104.12
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB
|
|
chr3_+_132389867
Show fit
|
0.23 |
ENSMUST00000169172.5
|
Tbck
|
TBC1 domain containing kinase
|
|
chr7_+_139616298
Show fit
|
0.23 |
ENSMUST00000168194.3
ENSMUST00000210882.2
|
Zfp511
|
zinc finger protein 511
|
|
chr5_-_51711237
Show fit
|
0.22 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr15_-_82796308
Show fit
|
0.22 |
ENSMUST00000109510.10
ENSMUST00000048966.7
|
Tcf20
|
transcription factor 20
|
|
chr2_+_156681991
Show fit
|
0.22 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2
|
|
chr10_-_61288437
Show fit
|
0.22 |
ENSMUST00000167087.2
ENSMUST00000020288.15
|
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr2_+_172314433
Show fit
|
0.22 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209
|
|
chr7_-_101859379
Show fit
|
0.22 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98
|
|
chr1_+_170136372
Show fit
|
0.22 |
ENSMUST00000056991.6
|
Spata46
|
spermatogenesis associated 46
|
|
chr17_+_18372149
Show fit
|
0.22 |
ENSMUST00000169686.3
|
Vmn2r92
|
vomeronasal 2, receptor 92
|
|
chr8_-_23143422
Show fit
|
0.21 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta
|
|
chr1_-_170133901
Show fit
|
0.21 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694
|
|
chr10_+_79984097
Show fit
|
0.21 |
ENSMUST00000099492.10
ENSMUST00000042057.12
|
Midn
|
midnolin
|
|
chr11_+_83193495
Show fit
|
0.21 |
ENSMUST00000176430.8
ENSMUST00000065692.14
ENSMUST00000142680.2
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr16_-_23807602
Show fit
|
0.21 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6
|
|
chr12_+_33197692
Show fit
|
0.21 |
ENSMUST00000077456.13
ENSMUST00000110824.9
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr13_-_38221045
Show fit
|
0.21 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1
|
|
chr19_+_6413840
Show fit
|
0.21 |
ENSMUST00000113488.8
ENSMUST00000113487.8
|
Sf1
|
splicing factor 1
|