Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
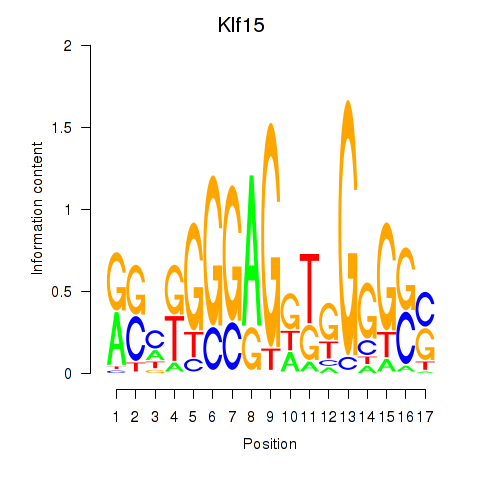
Results for Klf15
Z-value: 0.68
Transcription factors associated with Klf15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf15
|
ENSMUSG00000030087.12 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf15 | mm39_v1_chr6_+_90443293_90443382 | 0.95 | 1.2e-02 | Click! |
Activity profile of Klf15 motif
Sorted Z-values of Klf15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_174171979 | 0.55 |
ENSMUST00000109083.2
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_-_5719302 | 0.52 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_-_65650133 | 0.38 |
ENSMUST00000196121.2
|
Gm43552
|
predicted gene 43552 |
| chr18_+_61178211 | 0.32 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr2_-_90410922 | 0.30 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr18_+_82928782 | 0.29 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr5_+_135093056 | 0.29 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr2_-_155315708 | 0.27 |
ENSMUST00000109670.8
ENSMUST00000123293.8 |
Ncoa6
|
nuclear receptor coactivator 6 |
| chr11_-_101917745 | 0.26 |
ENSMUST00000107167.2
ENSMUST00000062801.11 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr13_-_58261406 | 0.26 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr17_+_34808772 | 0.26 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr7_-_19410749 | 0.25 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr6_-_124710030 | 0.25 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr15_-_82783978 | 0.24 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr9_-_43151179 | 0.22 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr9_+_21077010 | 0.21 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr2_-_38177359 | 0.21 |
ENSMUST00000102787.10
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr5_+_135197228 | 0.21 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chrX_-_135769285 | 0.21 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr10_-_80096842 | 0.21 |
ENSMUST00000105363.8
|
Gamt
|
guanidinoacetate methyltransferase |
| chr9_+_89791943 | 0.20 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr11_-_100541519 | 0.20 |
ENSMUST00000103119.10
|
Zfp385c
|
zinc finger protein 385C |
| chr4_+_123798690 | 0.20 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr15_+_81469538 | 0.20 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr17_+_3376827 | 0.20 |
ENSMUST00000169838.9
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr7_+_97229065 | 0.19 |
ENSMUST00000107153.3
|
Rsf1
|
remodeling and spacing factor 1 |
| chr13_+_38009981 | 0.19 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr7_+_127111576 | 0.19 |
ENSMUST00000186672.7
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr15_-_36609812 | 0.19 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_+_100355910 | 0.19 |
ENSMUST00000207875.2
ENSMUST00000208013.2 |
Fam168a
|
family with sequence similarity 168, member A |
| chr10_-_127190280 | 0.18 |
ENSMUST00000059718.6
|
Inhbe
|
inhibin beta-E |
| chr2_+_174171860 | 0.18 |
ENSMUST00000109087.8
ENSMUST00000109084.8 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_+_174171799 | 0.17 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr11_+_77873535 | 0.17 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr5_-_124563611 | 0.17 |
ENSMUST00000198420.5
|
Sbno1
|
strawberry notch 1 |
| chr12_+_102249375 | 0.17 |
ENSMUST00000101114.11
ENSMUST00000150795.8 |
Rin3
|
Ras and Rab interactor 3 |
| chr9_+_40597297 | 0.17 |
ENSMUST00000034522.8
|
Clmp
|
CXADR-like membrane protein |
| chr1_-_64161415 | 0.16 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_+_29759495 | 0.16 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr10_-_81262948 | 0.15 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr17_+_34341766 | 0.15 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr2_+_18069375 | 0.15 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr4_+_150699670 | 0.15 |
ENSMUST00000219467.2
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr5_+_97145533 | 0.15 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr16_+_33504908 | 0.15 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr16_+_4412546 | 0.15 |
ENSMUST00000014447.13
|
Glis2
|
GLIS family zinc finger 2 |
| chr15_+_74827488 | 0.14 |
ENSMUST00000191436.7
ENSMUST00000191145.7 |
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr2_+_27776428 | 0.14 |
ENSMUST00000028280.14
|
Col5a1
|
collagen, type V, alpha 1 |
| chr2_-_38177182 | 0.14 |
ENSMUST00000130472.8
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr15_-_89064936 | 0.14 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr7_+_46045862 | 0.14 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr3_-_27764571 | 0.14 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr14_-_56021484 | 0.13 |
ENSMUST00000170223.9
ENSMUST00000002398.9 ENSMUST00000227031.2 |
Adcy4
|
adenylate cyclase 4 |
| chr1_+_55445033 | 0.13 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr11_-_116197478 | 0.13 |
ENSMUST00000126731.8
|
Exoc7
|
exocyst complex component 7 |
| chr4_-_107035479 | 0.13 |
ENSMUST00000058585.14
|
Tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr4_+_47353217 | 0.13 |
ENSMUST00000007757.15
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr1_-_156301821 | 0.13 |
ENSMUST00000188027.2
ENSMUST00000187507.7 ENSMUST00000189661.7 |
Soat1
|
sterol O-acyltransferase 1 |
| chr18_-_89787603 | 0.12 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr5_+_135197137 | 0.12 |
ENSMUST00000031692.12
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr18_-_73836810 | 0.12 |
ENSMUST00000025393.14
|
Smad4
|
SMAD family member 4 |
| chr7_-_80052491 | 0.12 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_+_79984097 | 0.12 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr11_+_70453724 | 0.12 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr11_+_70453666 | 0.12 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr4_+_55350043 | 0.12 |
ENSMUST00000030134.9
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr2_+_164802766 | 0.12 |
ENSMUST00000202223.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr15_-_102165884 | 0.12 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr10_-_80096793 | 0.12 |
ENSMUST00000020359.7
|
Gamt
|
guanidinoacetate methyltransferase |
| chr8_-_89088782 | 0.12 |
ENSMUST00000034085.8
|
Brd7
|
bromodomain containing 7 |
| chr6_-_113354826 | 0.12 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr5_-_124563636 | 0.12 |
ENSMUST00000196711.5
ENSMUST00000200474.5 ENSMUST00000199808.5 |
Sbno1
|
strawberry notch 1 |
| chr4_+_119590971 | 0.11 |
ENSMUST00000084306.5
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr2_-_26335187 | 0.11 |
ENSMUST00000114082.9
ENSMUST00000091252.5 |
Sec16a
|
SEC16 homolog A, endoplasmic reticulum export factor |
| chr2_-_45002902 | 0.11 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_62067026 | 0.11 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr17_+_86475205 | 0.11 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr5_+_127709302 | 0.11 |
ENSMUST00000118139.3
|
Glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr3_+_135144287 | 0.11 |
ENSMUST00000196591.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_+_130477642 | 0.11 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr5_+_30745447 | 0.11 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr11_-_53959790 | 0.11 |
ENSMUST00000018755.10
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr5_-_86320599 | 0.11 |
ENSMUST00000039373.14
|
Uba6
|
ubiquitin-like modifier activating enzyme 6 |
| chr18_+_82928959 | 0.10 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr8_+_108020132 | 0.10 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr17_+_27152012 | 0.10 |
ENSMUST00000073724.7
ENSMUST00000237189.2 |
Phf1
|
PHD finger protein 1 |
| chr12_+_71062733 | 0.10 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr15_-_68130636 | 0.10 |
ENSMUST00000162173.8
ENSMUST00000160248.8 ENSMUST00000162054.9 |
Zfat
|
zinc finger and AT hook domain containing |
| chr2_-_33321306 | 0.10 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr11_-_107685383 | 0.10 |
ENSMUST00000021066.4
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr11_-_4696778 | 0.10 |
ENSMUST00000009219.3
|
Cabp7
|
calcium binding protein 7 |
| chr6_+_54016543 | 0.10 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr8_-_73302068 | 0.09 |
ENSMUST00000058534.7
|
Med26
|
mediator complex subunit 26 |
| chr2_-_9883391 | 0.09 |
ENSMUST00000102976.4
|
Gata3
|
GATA binding protein 3 |
| chr17_-_34341544 | 0.09 |
ENSMUST00000025193.14
|
Brd2
|
bromodomain containing 2 |
| chr4_+_107035566 | 0.09 |
ENSMUST00000030361.11
|
Tmem59
|
transmembrane protein 59 |
| chr18_+_82929451 | 0.09 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr13_-_51947685 | 0.09 |
ENSMUST00000110040.9
ENSMUST00000021900.14 |
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr5_-_124564014 | 0.09 |
ENSMUST00000196329.5
ENSMUST00000196644.5 |
Sbno1
|
strawberry notch 1 |
| chr14_+_76082533 | 0.09 |
ENSMUST00000110894.9
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr11_-_86248395 | 0.09 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr7_+_79676095 | 0.09 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chrX_+_101048650 | 0.08 |
ENSMUST00000144753.2
|
Nhsl2
|
NHS-like 2 |
| chr11_+_70453806 | 0.08 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr15_-_99355623 | 0.08 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr15_-_102165740 | 0.08 |
ENSMUST00000135466.2
|
Rarg
|
retinoic acid receptor, gamma |
| chr15_+_102178975 | 0.08 |
ENSMUST00000181801.2
|
Gm9918
|
predicted gene 9918 |
| chr6_+_124986193 | 0.08 |
ENSMUST00000112428.8
|
Zfp384
|
zinc finger protein 384 |
| chr18_-_38068456 | 0.08 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr1_-_72576089 | 0.08 |
ENSMUST00000047786.6
|
Marchf4
|
membrane associated ring-CH-type finger 4 |
| chr14_+_76652369 | 0.08 |
ENSMUST00000110888.8
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr4_+_62537750 | 0.08 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr1_-_84816379 | 0.08 |
ENSMUST00000187818.2
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr15_-_64184485 | 0.07 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr10_-_80382611 | 0.07 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr1_-_160862364 | 0.07 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr7_+_126376099 | 0.07 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr15_-_79287747 | 0.07 |
ENSMUST00000074991.10
|
Tmem184b
|
transmembrane protein 184b |
| chr2_-_91013362 | 0.07 |
ENSMUST00000066420.12
|
Madd
|
MAP-kinase activating death domain |
| chr10_-_81037878 | 0.07 |
ENSMUST00000005069.8
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr14_-_71004019 | 0.07 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr4_+_123798625 | 0.07 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr7_-_16348862 | 0.07 |
ENSMUST00000171937.2
ENSMUST00000075845.11 |
Arhgap35
|
Rho GTPase activating protein 35 |
| chr4_+_47353277 | 0.07 |
ENSMUST00000044234.14
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr10_+_20188269 | 0.07 |
ENSMUST00000185800.7
|
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr4_-_136563154 | 0.06 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr11_+_94827050 | 0.06 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr10_-_127502541 | 0.06 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_-_81515865 | 0.06 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr8_+_94537460 | 0.06 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_-_24643919 | 0.06 |
ENSMUST00000206705.3
|
Erfl
|
ETS repressor factor like |
| chr1_-_64160557 | 0.06 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr15_-_36609208 | 0.06 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_-_59549243 | 0.06 |
ENSMUST00000173699.8
ENSMUST00000173884.8 ENSMUST00000102883.11 ENSMUST00000174586.8 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr18_-_3337467 | 0.06 |
ENSMUST00000154135.8
ENSMUST00000142690.2 ENSMUST00000025069.11 ENSMUST00000165086.8 ENSMUST00000082141.12 ENSMUST00000149803.8 |
Crem
|
cAMP responsive element modulator |
| chr4_+_139107911 | 0.06 |
ENSMUST00000165860.8
ENSMUST00000097822.10 |
Ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr18_-_38068430 | 0.06 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1 |
| chr9_-_45817666 | 0.06 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr12_+_16944896 | 0.06 |
ENSMUST00000020904.8
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2 |
| chr7_+_100355798 | 0.06 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr8_+_12807001 | 0.06 |
ENSMUST00000033818.10
ENSMUST00000091237.12 |
Atp11a
|
ATPase, class VI, type 11A |
| chr2_+_38229270 | 0.06 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2 |
| chr12_+_71877838 | 0.06 |
ENSMUST00000223272.2
ENSMUST00000085299.4 |
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr7_-_110213968 | 0.06 |
ENSMUST00000166020.8
ENSMUST00000171218.8 ENSMUST00000033058.14 ENSMUST00000164759.8 |
Sbf2
|
SET binding factor 2 |
| chr17_+_29768757 | 0.06 |
ENSMUST00000048677.9
ENSMUST00000150388.3 |
Tbc1d22b
Gm28052
|
TBC1 domain family, member 22B predicted gene, 28052 |
| chr16_+_35361635 | 0.05 |
ENSMUST00000120756.8
|
Sema5b
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr10_-_18619439 | 0.05 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr12_-_4891435 | 0.05 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr9_-_44792575 | 0.05 |
ENSMUST00000114689.8
ENSMUST00000002095.11 ENSMUST00000128768.3 |
Kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr9_+_118307250 | 0.05 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr13_-_46118433 | 0.05 |
ENSMUST00000167708.4
ENSMUST00000091628.11 ENSMUST00000180110.9 |
Atxn1
|
ataxin 1 |
| chr2_+_140237229 | 0.05 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr7_-_116042674 | 0.05 |
ENSMUST00000170430.3
ENSMUST00000206219.2 |
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
| chr10_-_127098932 | 0.05 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr10_-_127502467 | 0.05 |
ENSMUST00000099157.4
|
Nab2
|
Ngfi-A binding protein 2 |
| chrX_-_151151680 | 0.05 |
ENSMUST00000070316.12
|
Gpr173
|
G-protein coupled receptor 173 |
| chr8_-_41586713 | 0.05 |
ENSMUST00000155055.2
ENSMUST00000059115.13 ENSMUST00000145860.2 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr4_+_47091909 | 0.05 |
ENSMUST00000045041.12
ENSMUST00000107744.2 |
Galnt12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr15_-_83609127 | 0.05 |
ENSMUST00000171496.9
ENSMUST00000043634.12 ENSMUST00000076060.12 ENSMUST00000016907.8 |
Scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr14_+_77394173 | 0.04 |
ENSMUST00000022589.9
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr2_-_44817218 | 0.04 |
ENSMUST00000100127.9
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr11_-_115310743 | 0.04 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr6_+_54017063 | 0.04 |
ENSMUST00000127323.3
|
Chn2
|
chimerin 2 |
| chr6_-_53045546 | 0.04 |
ENSMUST00000074541.6
|
Jazf1
|
JAZF zinc finger 1 |
| chr17_+_27152276 | 0.04 |
ENSMUST00000237412.2
|
Phf1
|
PHD finger protein 1 |
| chr16_-_13977084 | 0.04 |
ENSMUST00000090300.6
|
Marf1
|
meiosis regulator and mRNA stability 1 |
| chr11_-_52173391 | 0.04 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr17_+_43978377 | 0.04 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr10_-_117212826 | 0.04 |
ENSMUST00000177145.8
ENSMUST00000176670.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr19_-_44017637 | 0.04 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr2_-_20973692 | 0.04 |
ENSMUST00000114594.8
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr10_-_78427721 | 0.04 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr7_+_126028225 | 0.04 |
ENSMUST00000106407.9
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr3_+_22130779 | 0.04 |
ENSMUST00000193734.6
|
Tbl1xr1
|
transducin (beta)-like 1X-linked receptor 1 |
| chr17_-_11059172 | 0.04 |
ENSMUST00000041463.7
|
Pacrg
|
PARK2 co-regulated |
| chr16_+_33504829 | 0.04 |
ENSMUST00000152782.8
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr12_+_87313472 | 0.04 |
ENSMUST00000021425.8
|
Ahsa1
|
AHA1, activator of heat shock protein ATPase 1 |
| chr4_+_131649001 | 0.03 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr12_-_70158739 | 0.03 |
ENSMUST00000222835.2
ENSMUST00000221275.2 ENSMUST00000095666.13 |
Nin
|
ninein |
| chr19_+_10611574 | 0.03 |
ENSMUST00000055115.9
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr10_-_84938350 | 0.03 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr10_+_20188207 | 0.03 |
ENSMUST00000092678.10
ENSMUST00000043881.12 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr17_+_34809132 | 0.03 |
ENSMUST00000173772.2
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr6_+_113054592 | 0.03 |
ENSMUST00000113157.8
|
Setd5
|
SET domain containing 5 |
| chr17_+_43978280 | 0.03 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr1_+_75522902 | 0.03 |
ENSMUST00000124341.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr12_+_102094977 | 0.03 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr16_+_21023505 | 0.03 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr6_+_124986627 | 0.03 |
ENSMUST00000046064.17
ENSMUST00000152752.8 ENSMUST00000088308.10 ENSMUST00000112425.8 ENSMUST00000084275.12 |
Zfp384
|
zinc finger protein 384 |
| chr2_+_151544086 | 0.03 |
ENSMUST00000109872.2
|
Tmem74b
|
transmembrane protein 74B |
| chr2_-_91013193 | 0.03 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chr19_+_5790918 | 0.03 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr7_+_79882609 | 0.03 |
ENSMUST00000062915.9
|
Gdpgp1
|
GDP-D-glucose phosphorylase 1 |
| chrX_+_169106356 | 0.03 |
ENSMUST00000178693.4
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr7_+_19725390 | 0.03 |
ENSMUST00000207805.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr3_+_94385602 | 0.03 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr3_+_94385661 | 0.03 |
ENSMUST00000200342.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr11_-_72686627 | 0.03 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr2_-_84545504 | 0.03 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr5_-_137246611 | 0.03 |
ENSMUST00000196391.5
|
Muc3a
|
mucin 3A, cell surface associated |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.3 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.1 | 0.2 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.2 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) brainstem development(GO:0003360) positive regulation of luteinizing hormone secretion(GO:0033686) nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.0 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.3 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.0 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0043199 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |