Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
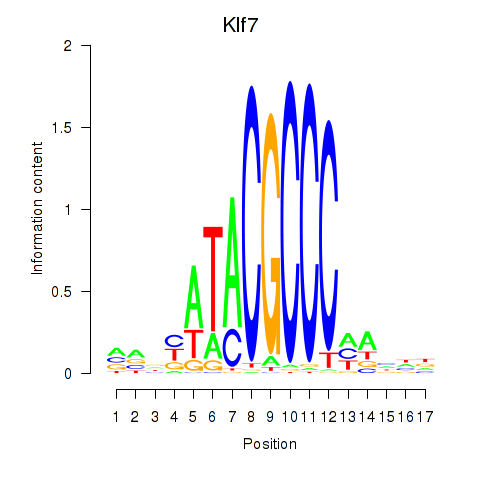
Results for Klf7
Z-value: 0.24
Transcription factors associated with Klf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf7
|
ENSMUSG00000025959.14 | Kruppel-like factor 7 (ubiquitous) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf7 | mm39_v1_chr1_-_64161415_64161455 | 0.36 | 5.5e-01 | Click! |
Activity profile of Klf7 motif
Sorted Z-values of Klf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_36227869 | 0.26 |
ENSMUST00000151664.8
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr11_-_115977755 | 0.19 |
ENSMUST00000074628.13
ENSMUST00000106444.4 |
Wbp2
|
WW domain binding protein 2 |
| chr15_-_34443054 | 0.17 |
ENSMUST00000142643.2
|
Rpl30
|
ribosomal protein L30 |
| chr5_-_115487005 | 0.12 |
ENSMUST00000040154.9
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr18_+_35252470 | 0.10 |
ENSMUST00000237154.2
|
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr9_+_103879745 | 0.10 |
ENSMUST00000035167.15
ENSMUST00000194774.6 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr3_-_103698391 | 0.10 |
ENSMUST00000106845.9
ENSMUST00000029438.15 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr1_-_138775317 | 0.10 |
ENSMUST00000093486.10
ENSMUST00000046870.13 |
Lhx9
|
LIM homeobox protein 9 |
| chr7_-_28465870 | 0.10 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr7_+_28466160 | 0.10 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr7_+_44804068 | 0.10 |
ENSMUST00000209954.2
|
Pih1d1
|
PIH1 domain containing 1 |
| chr2_-_156154667 | 0.09 |
ENSMUST00000079125.8
|
Scand1
|
SCAN domain-containing 1 |
| chr2_-_151815307 | 0.09 |
ENSMUST00000109863.2
|
Fam110a
|
family with sequence similarity 110, member A |
| chr11_+_78427181 | 0.09 |
ENSMUST00000128788.8
|
Ift20
|
intraflagellar transport 20 |
| chr17_+_36227433 | 0.09 |
ENSMUST00000087211.9
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr7_+_44803721 | 0.09 |
ENSMUST00000210139.2
ENSMUST00000211709.2 ENSMUST00000085375.13 ENSMUST00000209847.2 |
Pih1d1
|
PIH1 domain containing 1 |
| chr2_+_158217558 | 0.08 |
ENSMUST00000109488.8
|
Snhg11
|
small nucleolar RNA host gene 11 |
| chr15_-_102630589 | 0.08 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr5_+_137777111 | 0.08 |
ENSMUST00000126126.8
ENSMUST00000031739.6 |
Ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr11_+_68322945 | 0.08 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr2_-_160714904 | 0.07 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr1_+_58484669 | 0.07 |
ENSMUST00000151272.2
|
Nif3l1
|
Ngg1 interacting factor 3-like 1 (S. pombe) |
| chr15_-_98851423 | 0.07 |
ENSMUST00000134214.3
|
Gm49450
|
predicted gene, 49450 |
| chr9_-_32255604 | 0.07 |
ENSMUST00000034533.7
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr15_-_50752437 | 0.07 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr9_+_44151962 | 0.06 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr10_+_59057767 | 0.06 |
ENSMUST00000182161.2
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr9_-_88320937 | 0.06 |
ENSMUST00000173011.9
|
Snx14
|
sorting nexin 14 |
| chr2_-_73722874 | 0.06 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chr5_-_124325213 | 0.06 |
ENSMUST00000161938.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr11_-_93846453 | 0.06 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr19_-_4109446 | 0.05 |
ENSMUST00000189808.7
|
Gstp3
|
glutathione S-transferase pi 3 |
| chr1_+_63216281 | 0.05 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_-_120048788 | 0.05 |
ENSMUST00000027634.13
|
Dbi
|
diazepam binding inhibitor |
| chr16_-_91728531 | 0.04 |
ENSMUST00000023677.10
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr8_+_106587268 | 0.04 |
ENSMUST00000212610.2
ENSMUST00000212484.2 ENSMUST00000212200.2 |
Nutf2
|
nuclear transport factor 2 |
| chr8_+_3671599 | 0.04 |
ENSMUST00000207389.2
|
Pet100
|
PET100 homolog |
| chr2_-_73722932 | 0.04 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr10_-_128579879 | 0.04 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr12_+_49429574 | 0.04 |
ENSMUST00000179669.3
|
Foxg1
|
forkhead box G1 |
| chr1_+_120048890 | 0.04 |
ENSMUST00000027637.13
ENSMUST00000112644.9 ENSMUST00000056038.15 |
3110009E18Rik
|
RIKEN cDNA 3110009E18 gene |
| chr2_-_32178034 | 0.04 |
ENSMUST00000183946.8
ENSMUST00000113400.3 ENSMUST00000050410.11 |
Swi5
|
SWI5 recombination repair homolog (yeast) |
| chr7_+_79910948 | 0.03 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr7_-_139558599 | 0.03 |
ENSMUST00000209335.2
ENSMUST00000210254.2 ENSMUST00000121412.2 ENSMUST00000097970.9 |
Spef1l
|
sperm flagellar 1 like |
| chr11_+_115294560 | 0.03 |
ENSMUST00000153983.8
ENSMUST00000106539.10 ENSMUST00000103036.5 |
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr8_+_71849497 | 0.03 |
ENSMUST00000002473.10
|
Babam1
|
BRISC and BRCA1 A complex member 1 |
| chr1_-_120048667 | 0.03 |
ENSMUST00000151708.3
|
Dbi
|
diazepam binding inhibitor |
| chr3_+_106628987 | 0.03 |
ENSMUST00000130105.2
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr1_+_171156568 | 0.03 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr7_-_68398917 | 0.03 |
ENSMUST00000118110.3
|
Arrdc4
|
arrestin domain containing 4 |
| chr11_+_80274105 | 0.03 |
ENSMUST00000165565.8
ENSMUST00000188489.7 ENSMUST00000017567.14 ENSMUST00000108216.8 ENSMUST00000053740.15 |
Zfp207
|
zinc finger protein 207 |
| chr17_-_84773544 | 0.03 |
ENSMUST00000047524.10
|
Thada
|
thyroid adenoma associated |
| chr13_-_14787602 | 0.03 |
ENSMUST00000220621.2
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr12_-_11486544 | 0.03 |
ENSMUST00000072299.7
|
Vsnl1
|
visinin-like 1 |
| chr2_-_160714749 | 0.03 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr11_-_61384998 | 0.03 |
ENSMUST00000101085.9
ENSMUST00000079080.13 ENSMUST00000108714.2 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr2_+_156154508 | 0.02 |
ENSMUST00000073942.12
ENSMUST00000109580.2 |
Cnbd2
|
cyclic nucleotide binding domain containing 2 |
| chr17_-_46513499 | 0.02 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr14_+_30201569 | 0.02 |
ENSMUST00000022535.9
ENSMUST00000223658.2 |
Dcp1a
|
decapping mRNA 1A |
| chr9_+_70450130 | 0.02 |
ENSMUST00000049263.9
|
Sltm
|
SAFB-like, transcription modulator |
| chr18_-_38068456 | 0.02 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr1_-_23422230 | 0.02 |
ENSMUST00000027343.6
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr16_-_56537650 | 0.02 |
ENSMUST00000128551.8
|
Tfg
|
Trk-fused gene |
| chr7_-_109215960 | 0.02 |
ENSMUST00000077909.9
|
Denn2b
|
DENN domain containing 2B |
| chr8_+_106587212 | 0.02 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr11_+_78427219 | 0.02 |
ENSMUST00000050366.15
ENSMUST00000108275.2 |
Ift20
|
intraflagellar transport 20 |
| chr9_+_70450024 | 0.02 |
ENSMUST00000216816.2
|
Sltm
|
SAFB-like, transcription modulator |
| chr11_+_78067537 | 0.02 |
ENSMUST00000098545.12
|
Tlcd1
|
TLC domain containing 1 |
| chr3_+_106629209 | 0.02 |
ENSMUST00000106736.3
ENSMUST00000154973.2 ENSMUST00000098750.5 ENSMUST00000150513.2 |
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr18_-_38068430 | 0.01 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1 |
| chr4_+_127019826 | 0.01 |
ENSMUST00000094712.5
|
Tmem35b
|
transmembrane protein 35B |
| chr11_-_60618022 | 0.01 |
ENSMUST00000002889.5
|
Flii
|
flightless I actin binding protein |
| chr11_-_97774945 | 0.01 |
ENSMUST00000093939.4
|
Fbxo47
|
F-box protein 47 |
| chr17_-_13981626 | 0.01 |
ENSMUST00000135850.8
ENSMUST00000143162.8 ENSMUST00000130033.8 ENSMUST00000053376.12 |
Gm49936
Tcte2
|
predicted gene, 49936 t-complex-associated testis expressed 2 |
| chr2_-_164932670 | 0.01 |
ENSMUST00000109304.2
|
1700025C18Rik
|
RIKEN cDNA 1700025C18 gene |
| chr16_+_36514334 | 0.01 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr18_+_67266054 | 0.01 |
ENSMUST00000236771.2
ENSMUST00000237304.2 ENSMUST00000076605.9 |
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr15_+_98972850 | 0.01 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein |
| chr12_-_79343040 | 0.01 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr9_-_109105194 | 0.01 |
ENSMUST00000198048.5
|
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr9_-_71393175 | 0.01 |
ENSMUST00000233263.2
ENSMUST00000034720.12 |
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr10_+_86541675 | 0.01 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr7_-_44578834 | 0.01 |
ENSMUST00000107857.11
ENSMUST00000167930.8 ENSMUST00000085399.13 ENSMUST00000166972.9 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr18_-_9450097 | 0.01 |
ENSMUST00000053917.6
|
Ccny
|
cyclin Y |
| chr1_-_194774549 | 0.01 |
ENSMUST00000162650.8
|
Cd46
|
CD46 antigen, complement regulatory protein |
| chr11_+_11064125 | 0.01 |
ENSMUST00000109681.8
|
Vwc2
|
von Willebrand factor C domain containing 2 |
| chr7_+_25380263 | 0.01 |
ENSMUST00000205325.2
ENSMUST00000206913.2 |
B9d2
|
B9 protein domain 2 |
| chr8_+_85628557 | 0.01 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr6_+_83996344 | 0.01 |
ENSMUST00000204987.3
ENSMUST00000203695.3 ENSMUST00000204354.3 ENSMUST00000089595.12 ENSMUST00000113818.8 ENSMUST00000081904.7 |
Dysf
|
dysferlin |
| chr2_-_156022054 | 0.00 |
ENSMUST00000126992.8
ENSMUST00000146288.8 ENSMUST00000029149.13 ENSMUST00000109587.9 ENSMUST00000109584.8 |
Rbm39
|
RNA binding motif protein 39 |
| chr11_-_40624200 | 0.00 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr15_+_85619958 | 0.00 |
ENSMUST00000109422.8
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr9_+_46179899 | 0.00 |
ENSMUST00000121598.8
|
Apoa5
|
apolipoprotein A-V |
| chr9_-_32255533 | 0.00 |
ENSMUST00000216033.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr1_-_135846937 | 0.00 |
ENSMUST00000027667.13
|
Pkp1
|
plakophilin 1 |
| chr8_+_70605404 | 0.00 |
ENSMUST00000110146.9
ENSMUST00000110143.8 ENSMUST00000110141.9 ENSMUST00000110140.2 |
Mef2b
|
myocyte enhancer factor 2B |
| chr1_-_135846858 | 0.00 |
ENSMUST00000163260.8
|
Pkp1
|
plakophilin 1 |
| chr16_+_36514386 | 0.00 |
ENSMUST00000119464.2
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr1_+_92759324 | 0.00 |
ENSMUST00000045970.8
|
Gpc1
|
glypican 1 |
| chr15_-_102630496 | 0.00 |
ENSMUST00000171838.2
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr7_-_73663422 | 0.00 |
ENSMUST00000026896.10
ENSMUST00000191970.2 |
St8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr4_+_140737955 | 0.00 |
ENSMUST00000071977.9
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr15_+_85619765 | 0.00 |
ENSMUST00000109423.8
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr2_+_172392911 | 0.00 |
ENSMUST00000170744.2
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr7_+_27770655 | 0.00 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr1_+_173025375 | 0.00 |
ENSMUST00000216603.2
ENSMUST00000215844.2 |
Olfr218
|
olfactory receptor 218 |
| chr18_+_67266784 | 0.00 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chrX_+_135171002 | 0.00 |
ENSMUST00000178632.8
ENSMUST00000053540.11 |
Bex3
|
brain expressed X-linked 3 |
| chr9_-_32255556 | 0.00 |
ENSMUST00000214223.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr18_-_64688271 | 0.00 |
ENSMUST00000235459.2
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr3_+_51131868 | 0.00 |
ENSMUST00000023849.15
ENSMUST00000167780.2 |
Noct
|
nocturnin |
| chr19_-_11806388 | 0.00 |
ENSMUST00000061235.3
|
Olfr1417
|
olfactory receptor 1417 |
| chr1_+_57445487 | 0.00 |
ENSMUST00000027114.6
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr4_+_143120980 | 0.00 |
ENSMUST00000037419.4
|
Pramel1
|
PRAME like 1 |
| chr9_-_31824758 | 0.00 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr17_-_15129766 | 0.00 |
ENSMUST00000228233.2
|
Wdr27
|
WD repeat domain 27 |
| chr9_+_37683933 | 0.00 |
ENSMUST00000080634.4
|
Olfr875
|
olfactory receptor 875 |
| chr10_-_114638202 | 0.00 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chrX_+_135171049 | 0.00 |
ENSMUST00000113112.2
|
Bex3
|
brain expressed X-linked 3 |
| chr15_+_85620308 | 0.00 |
ENSMUST00000057979.6
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr11_+_63023893 | 0.00 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr11_+_42310557 | 0.00 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr4_-_156077061 | 0.00 |
ENSMUST00000052185.5
|
B3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase, polypeptide 6 |
| chr10_+_128030500 | 0.00 |
ENSMUST00000123291.2
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr5_-_3646071 | 0.00 |
ENSMUST00000121877.3
|
Rbm48
|
RNA binding motif protein 48 |
| chr4_-_87148672 | 0.00 |
ENSMUST00000107157.9
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |