Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
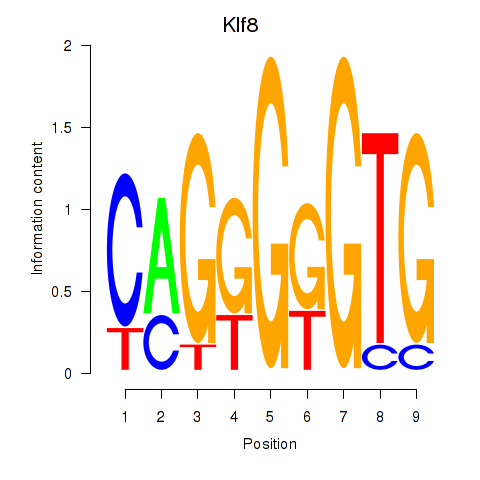
Results for Klf8
Z-value: 1.07
Transcription factors associated with Klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf8
|
ENSMUSG00000041649.14 | Kruppel-like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | mm39_v1_chrX_+_152020744_152020838 | 0.83 | 8.4e-02 | Click! |
Activity profile of Klf8 motif
Sorted Z-values of Klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_120151982 | 0.61 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr2_-_164197987 | 0.60 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr17_-_34219225 | 0.57 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr19_+_6952580 | 0.53 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr7_-_103477126 | 0.52 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr10_+_79716876 | 0.51 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr16_+_30418535 | 0.49 |
ENSMUST00000059078.4
|
Fam43a
|
family with sequence similarity 43, member A |
| chr19_-_6117815 | 0.48 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr16_+_17884267 | 0.46 |
ENSMUST00000151266.8
ENSMUST00000066027.14 ENSMUST00000155387.8 |
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr18_-_43610829 | 0.44 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr15_-_79976016 | 0.44 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr14_-_55828511 | 0.44 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr19_+_6952319 | 0.44 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr8_+_73449907 | 0.43 |
ENSMUST00000109950.5
ENSMUST00000004494.16 ENSMUST00000212095.2 ENSMUST00000212096.2 |
Sin3b
|
transcriptional regulator, SIN3B (yeast) |
| chr13_+_73476629 | 0.41 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr7_-_126625657 | 0.39 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_-_26014613 | 0.39 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr10_+_39245746 | 0.39 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr17_+_35117905 | 0.38 |
ENSMUST00000097342.10
ENSMUST00000013931.12 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr11_+_101215889 | 0.38 |
ENSMUST00000041095.14
ENSMUST00000107264.2 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr11_+_83637766 | 0.37 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr3_-_121608859 | 0.37 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_+_174169351 | 0.37 |
ENSMUST00000124935.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_-_164198427 | 0.36 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr14_+_122771734 | 0.36 |
ENSMUST00000154206.8
ENSMUST00000038374.13 ENSMUST00000135578.8 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chrX_-_74460168 | 0.34 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr17_+_46957151 | 0.33 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr17_+_35117438 | 0.33 |
ENSMUST00000114033.9
ENSMUST00000078061.13 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr11_-_120538928 | 0.32 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr1_-_188740023 | 0.32 |
ENSMUST00000085678.8
|
Kctd3
|
potassium channel tetramerisation domain containing 3 |
| chr8_+_86026318 | 0.32 |
ENSMUST00000170141.3
ENSMUST00000034132.13 |
Orc6
|
origin recognition complex, subunit 6 |
| chr17_+_74645936 | 0.31 |
ENSMUST00000224711.2
ENSMUST00000024869.8 ENSMUST00000233611.2 |
Spast
|
spastin |
| chr19_+_36325683 | 0.31 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr2_-_157408239 | 0.31 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr15_-_89258012 | 0.31 |
ENSMUST00000167643.4
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_-_6899173 | 0.30 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr2_-_28730286 | 0.30 |
ENSMUST00000037117.6
ENSMUST00000171404.8 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr3_-_121608809 | 0.29 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr12_+_28725218 | 0.29 |
ENSMUST00000020957.13
|
Adi1
|
acireductone dioxygenase 1 |
| chr7_-_126625617 | 0.29 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_+_122347792 | 0.29 |
ENSMUST00000072602.14
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr5_+_31078911 | 0.29 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr1_-_106641940 | 0.29 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr19_+_5540591 | 0.28 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle |
| chrX_+_7750483 | 0.28 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr5_+_115373895 | 0.28 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr5_+_139777263 | 0.27 |
ENSMUST00000018287.10
|
Mafk
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
| chr9_-_114610879 | 0.27 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr5_+_32616566 | 0.27 |
ENSMUST00000202078.2
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr4_-_150998857 | 0.26 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr10_-_80426125 | 0.26 |
ENSMUST00000187646.2
ENSMUST00000191440.7 ENSMUST00000003436.12 |
Abhd17a
|
abhydrolase domain containing 17A |
| chr10_-_90959817 | 0.26 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr11_+_115310885 | 0.25 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr1_-_66902429 | 0.25 |
ENSMUST00000027153.6
|
Acadl
|
acyl-Coenzyme A dehydrogenase, long-chain |
| chr8_-_85414220 | 0.25 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr11_-_101357046 | 0.25 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chrX_-_72965434 | 0.25 |
ENSMUST00000096316.4
ENSMUST00000114390.8 ENSMUST00000114391.10 ENSMUST00000114387.8 |
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr17_+_25992761 | 0.24 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr5_+_124045552 | 0.24 |
ENSMUST00000166233.2
|
Denr
|
density-regulated protein |
| chr5_-_122639840 | 0.24 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr9_-_96513529 | 0.24 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr19_-_6899121 | 0.24 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr8_+_86567600 | 0.24 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr17_-_26087696 | 0.23 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr19_-_12478803 | 0.23 |
ENSMUST00000045521.9
|
Dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr11_+_70548022 | 0.23 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr12_+_72807985 | 0.22 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr5_-_33812830 | 0.22 |
ENSMUST00000200849.2
|
Tmem129
|
transmembrane protein 129 |
| chr5_-_143255713 | 0.22 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr2_-_38534099 | 0.21 |
ENSMUST00000028083.6
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7 |
| chrX_-_74460137 | 0.21 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr8_-_86026385 | 0.21 |
ENSMUST00000034131.10
|
Vps35
|
VPS35 retromer complex component |
| chr19_+_45991907 | 0.21 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr5_+_31079177 | 0.21 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr11_+_115310963 | 0.20 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr15_-_97665524 | 0.20 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr2_+_121859025 | 0.20 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chrX_+_161543384 | 0.19 |
ENSMUST00000033720.12
ENSMUST00000112327.8 |
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chr11_+_60428788 | 0.18 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr1_+_135945798 | 0.18 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr15_-_89258034 | 0.18 |
ENSMUST00000228977.2
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr1_+_172327812 | 0.18 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chrX_-_84820209 | 0.18 |
ENSMUST00000142152.2
ENSMUST00000156390.8 |
Gk
|
glycerol kinase |
| chr8_+_22682816 | 0.18 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chrX_+_161543423 | 0.17 |
ENSMUST00000112326.8
|
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chr18_-_10610048 | 0.17 |
ENSMUST00000115864.8
ENSMUST00000145320.2 ENSMUST00000097670.10 |
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr4_+_148125630 | 0.17 |
ENSMUST00000069604.15
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chrX_-_72965524 | 0.17 |
ENSMUST00000114389.10
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr11_+_119833589 | 0.17 |
ENSMUST00000106231.8
ENSMUST00000075180.12 ENSMUST00000103021.10 ENSMUST00000026436.10 ENSMUST00000106233.2 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr7_-_142522995 | 0.17 |
ENSMUST00000009392.6
|
Ascl2
|
achaete-scute family bHLH transcription factor 2 |
| chr2_-_90735171 | 0.17 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr9_-_21202353 | 0.17 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr8_+_13389656 | 0.16 |
ENSMUST00000210165.2
ENSMUST00000170909.2 |
Tfdp1
|
transcription factor Dp 1 |
| chr14_-_68170873 | 0.16 |
ENSMUST00000039135.6
|
Dock5
|
dedicator of cytokinesis 5 |
| chr5_-_88823472 | 0.16 |
ENSMUST00000113234.8
ENSMUST00000153565.8 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr18_+_36414122 | 0.16 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr7_+_110628158 | 0.16 |
ENSMUST00000005749.6
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr1_-_20890437 | 0.16 |
ENSMUST00000053266.11
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chrX_-_72965536 | 0.16 |
ENSMUST00000033763.15
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr9_-_99450948 | 0.15 |
ENSMUST00000035043.12
|
Armc8
|
armadillo repeat containing 8 |
| chr14_-_20844074 | 0.15 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_+_167922924 | 0.15 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr4_+_155896946 | 0.15 |
ENSMUST00000030944.11
|
Ccnl2
|
cyclin L2 |
| chr2_+_90735077 | 0.15 |
ENSMUST00000111464.8
ENSMUST00000090682.4 |
Kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr11_-_45835737 | 0.15 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr3_+_96543143 | 0.15 |
ENSMUST00000165842.3
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr4_-_132649798 | 0.14 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr13_-_43324824 | 0.14 |
ENSMUST00000220787.2
|
Tbc1d7
|
TBC1 domain family, member 7 |
| chr9_-_45847344 | 0.14 |
ENSMUST00000034590.4
|
Tagln
|
transgelin |
| chr5_-_123270449 | 0.14 |
ENSMUST00000186469.7
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr5_+_122348140 | 0.14 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr4_-_149747644 | 0.14 |
ENSMUST00000105689.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr7_+_101070897 | 0.14 |
ENSMUST00000163751.10
ENSMUST00000211368.2 ENSMUST00000166652.2 |
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated |
| chr15_+_5173342 | 0.14 |
ENSMUST00000051186.9
ENSMUST00000228218.2 |
Prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr5_-_123270702 | 0.14 |
ENSMUST00000031401.6
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr4_+_115641996 | 0.13 |
ENSMUST00000177280.8
ENSMUST00000176047.8 |
Atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr9_-_21202693 | 0.13 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr1_+_135945705 | 0.13 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr17_-_71158184 | 0.13 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_-_120489172 | 0.13 |
ENSMUST00000026125.3
|
Alyref
|
Aly/REF export factor |
| chr17_+_29487881 | 0.13 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr11_+_86574811 | 0.13 |
ENSMUST00000108022.8
ENSMUST00000108021.2 |
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr8_-_86567506 | 0.13 |
ENSMUST00000034140.9
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr7_+_30264835 | 0.13 |
ENSMUST00000043850.14
|
Igflr1
|
IGF-like family receptor 1 |
| chr19_+_42078859 | 0.12 |
ENSMUST00000235932.2
ENSMUST00000066778.6 |
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr10_+_80003612 | 0.12 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr17_-_3608056 | 0.12 |
ENSMUST00000041003.8
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr12_-_69274936 | 0.12 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr5_+_129924564 | 0.12 |
ENSMUST00000041466.14
|
Zbed5
|
zinc finger, BED type containing 5 |
| chrX_-_84820250 | 0.12 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr7_+_28937859 | 0.11 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chrX_+_20524565 | 0.11 |
ENSMUST00000089217.11
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr9_+_64868127 | 0.11 |
ENSMUST00000170517.9
ENSMUST00000037504.7 |
Ints14
|
integrator complex subunit 14 |
| chr17_-_25179635 | 0.11 |
ENSMUST00000024981.9
|
Jpt2
|
Jupiter microtubule associated homolog 2 |
| chr10_-_90959853 | 0.11 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr17_-_71158052 | 0.11 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr9_+_108765701 | 0.11 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr11_+_120489358 | 0.10 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr17_+_47983587 | 0.10 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr2_+_122479770 | 0.10 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chrX_-_7834057 | 0.10 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr15_+_78290975 | 0.10 |
ENSMUST00000043865.8
ENSMUST00000231159.2 ENSMUST00000169133.8 |
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr4_+_118745806 | 0.10 |
ENSMUST00000106361.3
ENSMUST00000105035.3 |
Olfr1330
|
olfactory receptor 1330 |
| chr12_-_79219382 | 0.10 |
ENSMUST00000055262.13
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr7_-_19133783 | 0.09 |
ENSMUST00000047170.10
ENSMUST00000108459.9 |
Klc3
|
kinesin light chain 3 |
| chr18_+_21134928 | 0.09 |
ENSMUST00000234536.2
ENSMUST00000052396.7 |
Rnf138
|
ring finger protein 138 |
| chr7_-_92523396 | 0.09 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr7_-_6334239 | 0.09 |
ENSMUST00000127658.2
ENSMUST00000062765.14 |
Zfp583
|
zinc finger protein 583 |
| chr4_-_130068902 | 0.09 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr17_-_71158703 | 0.08 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr9_+_119766672 | 0.08 |
ENSMUST00000035100.6
|
Ttc21a
|
tetratricopeptide repeat domain 21A |
| chr10_-_5755412 | 0.08 |
ENSMUST00000019907.8
|
Fbxo5
|
F-box protein 5 |
| chr7_+_28937746 | 0.08 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr6_-_28397997 | 0.08 |
ENSMUST00000035930.11
|
Zfp800
|
zinc finger protein 800 |
| chrX_-_50294652 | 0.08 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr7_+_28937898 | 0.08 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_+_11634967 | 0.08 |
ENSMUST00000141436.8
ENSMUST00000126058.8 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr5_+_32616187 | 0.08 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr8_+_85763534 | 0.08 |
ENSMUST00000093360.12
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr11_-_76386190 | 0.07 |
ENSMUST00000108408.9
|
Abr
|
active BCR-related gene |
| chr5_+_88712840 | 0.07 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr19_-_12742811 | 0.07 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr15_-_97665679 | 0.07 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr9_-_106035308 | 0.07 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr5_-_108808649 | 0.07 |
ENSMUST00000053913.13
|
Dgkq
|
diacylglycerol kinase, theta |
| chr8_+_85763780 | 0.07 |
ENSMUST00000211601.2
ENSMUST00000166592.2 |
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr12_+_18564466 | 0.07 |
ENSMUST00000177778.9
ENSMUST00000063216.6 |
5730507C01Rik
|
RIKEN cDNA 5730507C01 gene |
| chr2_-_136958544 | 0.06 |
ENSMUST00000028735.8
|
Jag1
|
jagged 1 |
| chr11_-_76462353 | 0.06 |
ENSMUST00000072740.13
|
Abr
|
active BCR-related gene |
| chr16_-_94171533 | 0.06 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_+_105991280 | 0.06 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr8_-_71112295 | 0.06 |
ENSMUST00000211715.2
ENSMUST00000210307.2 ENSMUST00000209768.2 ENSMUST00000070173.9 |
Pgpep1
|
pyroglutamyl-peptidase I |
| chr7_+_51528788 | 0.06 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr12_-_118265103 | 0.06 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr11_+_70548513 | 0.06 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr15_-_83033559 | 0.06 |
ENSMUST00000058793.14
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr19_-_53026965 | 0.05 |
ENSMUST00000183274.8
ENSMUST00000182097.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr17_-_46956920 | 0.05 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chr11_-_62539284 | 0.05 |
ENSMUST00000057194.9
|
Lrrc75a
|
leucine rich repeat containing 75A |
| chr2_+_28403084 | 0.05 |
ENSMUST00000135803.8
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr2_+_32496957 | 0.05 |
ENSMUST00000113290.8
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chrX_+_74460275 | 0.05 |
ENSMUST00000118428.8
ENSMUST00000114074.8 ENSMUST00000133781.8 |
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr15_+_78290896 | 0.05 |
ENSMUST00000167140.8
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr9_+_37466989 | 0.05 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chr1_-_51517548 | 0.05 |
ENSMUST00000027279.12
ENSMUST00000186684.7 ENSMUST00000188204.7 ENSMUST00000185534.2 ENSMUST00000188051.7 |
Nabp1
|
nucleic acid binding protein 1 |
| chr16_-_18630365 | 0.04 |
ENSMUST00000096990.10
|
Cdc45
|
cell division cycle 45 |
| chr2_+_174169492 | 0.04 |
ENSMUST00000156623.8
ENSMUST00000149016.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr10_+_4660119 | 0.04 |
ENSMUST00000105588.8
ENSMUST00000105589.2 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr9_-_34967081 | 0.04 |
ENSMUST00000215463.2
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_-_157072060 | 0.04 |
ENSMUST00000129880.8
|
Rasal2
|
RAS protein activator like 2 |
| chr2_-_91540864 | 0.04 |
ENSMUST00000028678.9
ENSMUST00000076803.12 |
Atg13
|
autophagy related 13 |
| chr8_+_95781722 | 0.04 |
ENSMUST00000058479.7
|
Drc7
|
dynein regulatory complex subunit 7 |
| chr7_+_16576821 | 0.03 |
ENSMUST00000168093.9
|
Prkd2
|
protein kinase D2 |
| chr9_-_21202545 | 0.03 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chrX_-_59449137 | 0.03 |
ENSMUST00000033480.13
ENSMUST00000101527.3 |
Atp11c
|
ATPase, class VI, type 11C |
| chr2_-_169973076 | 0.03 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr9_+_114807546 | 0.03 |
ENSMUST00000183104.8
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr14_-_47025724 | 0.03 |
ENSMUST00000146629.3
ENSMUST00000015903.12 |
Cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr7_-_132616977 | 0.03 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr14_+_54713557 | 0.03 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.7 | GO:0036166 | phenotypic switching(GO:0036166) cellular response to cocaine(GO:0071314) |
| 0.1 | 0.5 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.6 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.5 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.4 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 0.2 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.1 | 0.5 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.6 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.3 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 1.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.6 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.1 | 0.2 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 0.4 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 0.3 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0044218 | growing cell tip(GO:0035838) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.5 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.2 | 0.5 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.1 | 0.4 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.4 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |