Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
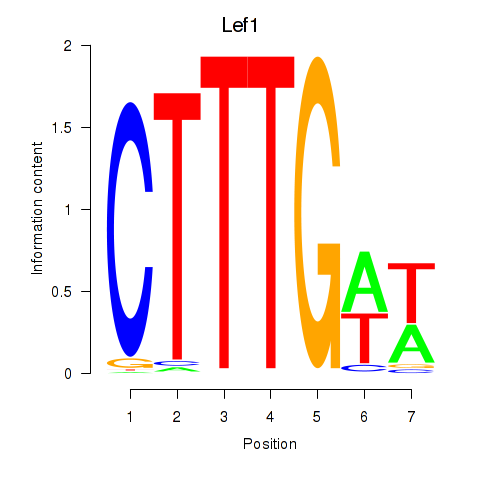
Results for Lef1
Z-value: 0.96
Transcription factors associated with Lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lef1
|
ENSMUSG00000027985.15 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | mm39_v1_chr3_+_130906434_130906452 | 0.79 | 1.1e-01 | Click! |
Activity profile of Lef1 motif
Sorted Z-values of Lef1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_18053158 | 1.04 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr14_-_98406977 | 1.01 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr19_-_30526916 | 0.70 |
ENSMUST00000025803.9
|
Dkk1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr9_-_49710058 | 0.64 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_-_48747232 | 0.62 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr13_-_54835460 | 0.61 |
ENSMUST00000129881.8
|
Rnf44
|
ring finger protein 44 |
| chr15_-_58078274 | 0.58 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr10_-_95159933 | 0.55 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr6_-_99073156 | 0.51 |
ENSMUST00000175886.8
|
Foxp1
|
forkhead box P1 |
| chr9_-_49710190 | 0.51 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_-_48747474 | 0.48 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr2_-_34261121 | 0.48 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr7_+_48895879 | 0.46 |
ENSMUST00000064395.13
|
Nav2
|
neuron navigator 2 |
| chrX_-_133062677 | 0.46 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr1_-_140111138 | 0.46 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr8_+_109441276 | 0.45 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr1_-_140111018 | 0.39 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr2_-_18053595 | 0.39 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr6_-_99005835 | 0.38 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1 |
| chr6_-_52141796 | 0.36 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr10_-_12745109 | 0.35 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr1_+_39940189 | 0.35 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_+_26895206 | 0.35 |
ENSMUST00000179391.8
ENSMUST00000108379.8 |
BC024978
|
cDNA sequence BC024978 |
| chrX_+_128650486 | 0.35 |
ENSMUST00000167619.9
ENSMUST00000037854.15 |
Diaph2
|
diaphanous related formin 2 |
| chr11_+_108811168 | 0.34 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr10_+_85763545 | 0.34 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr17_-_32503107 | 0.33 |
ENSMUST00000237692.2
|
Brd4
|
bromodomain containing 4 |
| chr8_-_85526653 | 0.33 |
ENSMUST00000126806.2
ENSMUST00000076715.13 |
Nfix
|
nuclear factor I/X |
| chr11_+_87959067 | 0.33 |
ENSMUST00000018521.11
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr1_-_135615831 | 0.33 |
ENSMUST00000190298.8
|
Nav1
|
neuron navigator 1 |
| chr9_+_119978773 | 0.32 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr13_-_54835508 | 0.32 |
ENSMUST00000177950.8
ENSMUST00000146931.8 |
Rnf44
|
ring finger protein 44 |
| chr7_+_27878894 | 0.31 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr3_+_53371086 | 0.31 |
ENSMUST00000058577.5
|
Proser1
|
proline and serine rich 1 |
| chr2_+_90912710 | 0.29 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr4_-_63321591 | 0.29 |
ENSMUST00000035724.5
|
Akna
|
AT-hook transcription factor |
| chrX_+_158623460 | 0.27 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr17_-_32503060 | 0.27 |
ENSMUST00000003726.16
ENSMUST00000121285.8 ENSMUST00000120276.9 |
Brd4
|
bromodomain containing 4 |
| chr12_+_85043262 | 0.27 |
ENSMUST00000101202.10
|
Ylpm1
|
YLP motif containing 1 |
| chr15_-_50753437 | 0.26 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr16_+_32090286 | 0.26 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr7_-_26895561 | 0.25 |
ENSMUST00000122202.8
ENSMUST00000080356.10 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr13_+_38010879 | 0.25 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chrX_+_128650579 | 0.25 |
ENSMUST00000113320.3
|
Diaph2
|
diaphanous related formin 2 |
| chr4_-_123558516 | 0.25 |
ENSMUST00000147030.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_-_156154667 | 0.25 |
ENSMUST00000079125.8
|
Scand1
|
SCAN domain-containing 1 |
| chr16_-_89940652 | 0.25 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr5_+_150119860 | 0.24 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr15_-_66432938 | 0.24 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr2_-_27365612 | 0.24 |
ENSMUST00000147736.2
|
Brd3
|
bromodomain containing 3 |
| chrX_+_158491589 | 0.24 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr12_+_76593799 | 0.24 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_+_166828982 | 0.24 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr14_+_47536075 | 0.24 |
ENSMUST00000227554.2
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr2_-_69416365 | 0.24 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr8_+_89247976 | 0.23 |
ENSMUST00000034086.13
|
Nkd1
|
naked cuticle 1 |
| chr8_+_66419809 | 0.23 |
ENSMUST00000072482.13
|
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr5_+_16758538 | 0.23 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr2_-_60503998 | 0.23 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr12_+_85043083 | 0.23 |
ENSMUST00000168977.8
ENSMUST00000021670.15 |
Ylpm1
|
YLP motif containing 1 |
| chr11_-_77380492 | 0.23 |
ENSMUST00000037593.14
ENSMUST00000092892.10 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr8_+_108669276 | 0.23 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr15_+_102885467 | 0.23 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr4_-_123421441 | 0.22 |
ENSMUST00000147228.8
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chrX_+_152281200 | 0.22 |
ENSMUST00000060714.10
|
Ubqln2
|
ubiquilin 2 |
| chr7_+_27879650 | 0.22 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr3_-_75864195 | 0.22 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr16_-_4831349 | 0.22 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr15_-_50753061 | 0.22 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr15_+_81350538 | 0.21 |
ENSMUST00000230219.2
|
Rbx1
|
ring-box 1 |
| chr5_-_132570710 | 0.21 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr8_+_40964818 | 0.21 |
ENSMUST00000098817.4
|
Vps37a
|
vacuolar protein sorting 37A |
| chr5_+_16758777 | 0.21 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr1_+_55445033 | 0.21 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr8_+_85807566 | 0.21 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr5_+_53748323 | 0.21 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr17_+_26633794 | 0.20 |
ENSMUST00000182897.8
ENSMUST00000183077.8 ENSMUST00000053020.8 |
Neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr9_-_51076724 | 0.20 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr18_+_82932747 | 0.20 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr12_-_84265609 | 0.20 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr8_+_85807369 | 0.19 |
ENSMUST00000079764.14
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr1_+_136059101 | 0.19 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr1_+_132119169 | 0.19 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr6_-_52211882 | 0.19 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr5_-_136596094 | 0.19 |
ENSMUST00000175975.9
ENSMUST00000176216.9 ENSMUST00000176745.8 |
Cux1
|
cut-like homeobox 1 |
| chr11_-_104332528 | 0.19 |
ENSMUST00000106971.2
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr17_+_74835290 | 0.19 |
ENSMUST00000180037.8
|
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr9_+_72832904 | 0.19 |
ENSMUST00000038489.6
|
Pygo1
|
pygopus 1 |
| chr5_+_77414031 | 0.19 |
ENSMUST00000113449.2
|
Rest
|
RE1-silencing transcription factor |
| chr15_-_98769056 | 0.19 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chrX_-_142716200 | 0.19 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr10_-_85847697 | 0.18 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr2_+_92015780 | 0.18 |
ENSMUST00000128781.9
ENSMUST00000111291.9 |
Phf21a
|
PHD finger protein 21A |
| chr4_+_116415251 | 0.18 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_-_29204812 | 0.18 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr8_-_11362731 | 0.18 |
ENSMUST00000033898.10
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr11_+_78006391 | 0.18 |
ENSMUST00000155571.2
|
Fam222b
|
family with sequence similarity 222, member B |
| chr3_+_130904000 | 0.18 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr16_+_17269845 | 0.18 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr10_+_66932235 | 0.18 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr10_-_60055082 | 0.18 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chrX_-_156198282 | 0.17 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr7_-_26895141 | 0.17 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr19_-_4384029 | 0.17 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr15_-_71906051 | 0.17 |
ENSMUST00000159993.8
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr12_+_71095112 | 0.16 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr9_+_61280764 | 0.16 |
ENSMUST00000160541.8
ENSMUST00000161207.8 ENSMUST00000159630.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr15_-_50753792 | 0.16 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chrX_+_101048650 | 0.16 |
ENSMUST00000144753.2
|
Nhsl2
|
NHS-like 2 |
| chr1_+_135074520 | 0.16 |
ENSMUST00000027684.11
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chr8_+_45960804 | 0.16 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_+_74835417 | 0.16 |
ENSMUST00000182944.8
ENSMUST00000182597.8 ENSMUST00000182133.8 ENSMUST00000183224.8 |
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr7_-_73187369 | 0.16 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr4_-_151946124 | 0.16 |
ENSMUST00000169423.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr14_+_58308004 | 0.16 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr18_+_4921663 | 0.16 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chr2_+_146063841 | 0.16 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1 |
| chrX_-_163041185 | 0.15 |
ENSMUST00000112265.9
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chrX_-_72868544 | 0.15 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr11_+_117223161 | 0.15 |
ENSMUST00000106349.2
|
Septin9
|
septin 9 |
| chr3_+_7568481 | 0.15 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr9_-_103639018 | 0.15 |
ENSMUST00000215136.2
ENSMUST00000189588.7 |
Tmem108
|
transmembrane protein 108 |
| chr9_-_103638985 | 0.15 |
ENSMUST00000189066.2
|
Tmem108
|
transmembrane protein 108 |
| chr10_+_79984097 | 0.15 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr19_-_14575395 | 0.15 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr2_+_18059982 | 0.14 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr8_+_88978600 | 0.14 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr11_+_79551358 | 0.14 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr19_-_5779648 | 0.14 |
ENSMUST00000116558.3
ENSMUST00000099955.4 ENSMUST00000161368.2 |
Fam89b
|
family with sequence similarity 89, member B |
| chr11_+_108814007 | 0.14 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr16_+_95058417 | 0.14 |
ENSMUST00000113861.8
ENSMUST00000113854.8 ENSMUST00000113862.8 ENSMUST00000037154.14 ENSMUST00000113855.8 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_131671751 | 0.13 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr1_-_156766351 | 0.13 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr11_+_6339442 | 0.13 |
ENSMUST00000109786.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr4_-_20778847 | 0.13 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr16_+_19916292 | 0.13 |
ENSMUST00000023509.5
ENSMUST00000232088.2 ENSMUST00000231842.2 |
Klhl24
|
kelch-like 24 |
| chr1_+_64572050 | 0.13 |
ENSMUST00000190348.2
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr6_+_17463819 | 0.13 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr6_+_15185202 | 0.13 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr8_+_95720864 | 0.13 |
ENSMUST00000212141.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr13_+_42205790 | 0.13 |
ENSMUST00000220525.2
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr13_+_89687915 | 0.13 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_-_65068960 | 0.12 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr5_-_71253107 | 0.12 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr8_+_95721378 | 0.12 |
ENSMUST00000212956.2
ENSMUST00000212531.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_+_107649900 | 0.12 |
ENSMUST00000214599.2
ENSMUST00000209805.3 |
Olfr479
|
olfactory receptor 479 |
| chr11_+_6339330 | 0.12 |
ENSMUST00000012612.11
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr14_-_36628263 | 0.12 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr6_-_97436223 | 0.12 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr9_+_61280890 | 0.12 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3 |
| chr13_+_83672965 | 0.12 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chrX_-_16777913 | 0.12 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr8_+_95721019 | 0.12 |
ENSMUST00000212976.2
ENSMUST00000212995.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr15_-_82796308 | 0.12 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr9_+_109961079 | 0.12 |
ENSMUST00000197480.5
ENSMUST00000197984.5 ENSMUST00000199896.2 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr19_+_8568618 | 0.12 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr4_-_151946155 | 0.11 |
ENSMUST00000049790.14
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr3_-_108117754 | 0.11 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr17_+_48717007 | 0.11 |
ENSMUST00000167180.8
ENSMUST00000046651.7 ENSMUST00000233426.2 |
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr17_-_33979280 | 0.11 |
ENSMUST00000173860.8
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr1_-_156766381 | 0.11 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chrX_-_37653396 | 0.11 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr1_+_42734051 | 0.11 |
ENSMUST00000239323.2
ENSMUST00000199521.5 ENSMUST00000176807.3 |
Pou3f3
Gm20646
|
POU domain, class 3, transcription factor 3 predicted gene 20646 |
| chr1_+_180468895 | 0.11 |
ENSMUST00000192725.6
|
Lin9
|
lin-9 homolog (C. elegans) |
| chr12_-_72283465 | 0.11 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr17_-_33979442 | 0.11 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr3_+_101917455 | 0.11 |
ENSMUST00000066187.6
ENSMUST00000198675.2 |
Nhlh2
|
nescient helix loop helix 2 |
| chr11_-_102815910 | 0.11 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr2_+_136555364 | 0.10 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr8_+_40964830 | 0.10 |
ENSMUST00000238339.2
|
Vps37a
|
vacuolar protein sorting 37A |
| chr7_-_78228116 | 0.10 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr5_+_77413282 | 0.10 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr9_+_109961048 | 0.10 |
ENSMUST00000088716.12
|
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr14_-_100521888 | 0.10 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
| chr15_+_81350497 | 0.10 |
ENSMUST00000023036.7
|
Rbx1
|
ring-box 1 |
| chr13_+_81031512 | 0.10 |
ENSMUST00000099356.10
|
Arrdc3
|
arrestin domain containing 3 |
| chr5_-_136596299 | 0.10 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr3_+_101917392 | 0.10 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr19_-_27988534 | 0.10 |
ENSMUST00000174850.8
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr15_-_50746202 | 0.10 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr2_-_170248421 | 0.10 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr18_+_65022035 | 0.10 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr9_+_21527462 | 0.10 |
ENSMUST00000034707.15
ENSMUST00000098948.10 |
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr13_+_35925296 | 0.10 |
ENSMUST00000163595.3
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr11_+_45946800 | 0.10 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr15_-_98851423 | 0.10 |
ENSMUST00000134214.3
|
Gm49450
|
predicted gene, 49450 |
| chr2_+_96148418 | 0.10 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr2_+_156154508 | 0.10 |
ENSMUST00000073942.12
ENSMUST00000109580.2 |
Cnbd2
|
cyclic nucleotide binding domain containing 2 |
| chr14_-_71004019 | 0.10 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr14_+_26761023 | 0.09 |
ENSMUST00000223942.2
|
Il17rd
|
interleukin 17 receptor D |
| chr5_-_51711237 | 0.09 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr6_+_54016543 | 0.09 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr4_-_141325517 | 0.09 |
ENSMUST00000131317.8
ENSMUST00000006381.11 ENSMUST00000129602.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr7_-_110681402 | 0.09 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr17_-_48716756 | 0.09 |
ENSMUST00000160319.8
ENSMUST00000159535.2 ENSMUST00000078800.13 ENSMUST00000046719.14 ENSMUST00000162460.8 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr11_-_52174129 | 0.09 |
ENSMUST00000109071.3
|
Tcf7
|
transcription factor 7, T cell specific |
| chr11_+_70323452 | 0.09 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr10_-_13264574 | 0.09 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_-_31763859 | 0.09 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chr2_+_172863688 | 0.09 |
ENSMUST00000029014.16
|
Rbm38
|
RNA binding motif protein 38 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lef1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.2 | 0.7 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.2 | 0.6 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 1.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.3 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.4 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.3 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.6 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.2 | GO:1905066 | Notch signaling pathway involved in heart induction(GO:0003137) regulation of Notch signaling pathway involved in heart induction(GO:0035480) positive regulation of Notch signaling pathway involved in heart induction(GO:0035481) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.0 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:1904633 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:1902661 | DNA methylation on cytosine within a CG sequence(GO:0010424) regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0060983 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.0 | GO:0032487 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.0 | 0.0 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 0.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |