Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
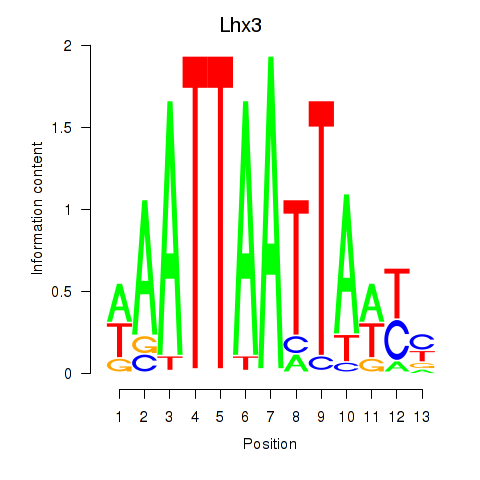
Results for Lhx3
Z-value: 0.24
Transcription factors associated with Lhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx3
|
ENSMUSG00000026934.16 | LIM homeobox protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx3 | mm39_v1_chr2_-_26096547_26096592 | -0.84 | 7.6e-02 | Click! |
Activity profile of Lhx3 motif
Sorted Z-values of Lhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_60907698 | 0.26 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr18_+_60907668 | 0.22 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr3_+_142326363 | 0.20 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr15_+_65682066 | 0.18 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr2_+_36120438 | 0.17 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr10_-_35587888 | 0.16 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr14_+_69585036 | 0.16 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr7_-_45480200 | 0.15 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr19_-_24178000 | 0.12 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chrM_+_9459 | 0.11 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr17_-_36343573 | 0.11 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr12_-_84664001 | 0.11 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr11_-_4045343 | 0.09 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr11_+_109434519 | 0.09 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr9_+_21634779 | 0.08 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr2_+_163500290 | 0.08 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr9_+_120758282 | 0.08 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr13_-_43634695 | 0.07 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chrX_+_41241049 | 0.07 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr1_+_177272297 | 0.07 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr12_-_25147139 | 0.07 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr2_-_84255602 | 0.07 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr3_+_151143524 | 0.06 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr7_+_99808452 | 0.06 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr5_+_122560501 | 0.06 |
ENSMUST00000031422.10
ENSMUST00000119792.2 |
Anapc7
|
anaphase promoting complex subunit 7 |
| chr5_+_96357337 | 0.06 |
ENSMUST00000117766.8
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr3_+_95226093 | 0.06 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr3_+_84859453 | 0.05 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr17_-_35081129 | 0.05 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr2_+_152468850 | 0.05 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr7_+_126575752 | 0.05 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr6_-_115014777 | 0.05 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr9_-_103569984 | 0.04 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr11_-_106205320 | 0.04 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr13_+_93441307 | 0.04 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chrX_-_8118541 | 0.04 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr1_+_177272215 | 0.04 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr17_-_35081456 | 0.04 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr13_+_93440572 | 0.04 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr10_-_62343516 | 0.04 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr10_-_37014859 | 0.04 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr17_-_56343531 | 0.03 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chrX_+_99669343 | 0.03 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr4_+_147216495 | 0.03 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr5_-_3697806 | 0.03 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr9_+_111014254 | 0.03 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr14_-_70945434 | 0.03 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr2_+_110427643 | 0.03 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chrX_+_65696608 | 0.03 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr11_+_6510167 | 0.03 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr9_-_110576124 | 0.03 |
ENSMUST00000199862.5
ENSMUST00000198865.5 |
Pth1r
|
parathyroid hormone 1 receptor |
| chr2_+_22959223 | 0.03 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr3_+_151143557 | 0.02 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr10_-_130265572 | 0.02 |
ENSMUST00000171811.4
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chr10_+_5543769 | 0.02 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr7_+_45271229 | 0.02 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr14_+_80237691 | 0.02 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr4_+_146695418 | 0.02 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr4_-_58499398 | 0.02 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr4_+_102446883 | 0.02 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr17_-_78991691 | 0.02 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr13_-_101829070 | 0.02 |
ENSMUST00000187009.7
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr14_+_53310220 | 0.02 |
ENSMUST00000196079.2
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chrX_-_99669507 | 0.02 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr2_+_22959452 | 0.02 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr11_-_30148230 | 0.02 |
ENSMUST00000102838.10
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr6_+_34757346 | 0.02 |
ENSMUST00000115016.8
ENSMUST00000115017.8 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr17_+_79919267 | 0.02 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr5_-_86437119 | 0.02 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr19_+_20579322 | 0.02 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr6_-_70313491 | 0.02 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr4_-_14621805 | 0.02 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr17_-_56343625 | 0.02 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr11_-_99265721 | 0.02 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr2_+_89642395 | 0.01 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr4_+_109092459 | 0.01 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr10_+_115979787 | 0.01 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr19_-_34143437 | 0.01 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr4_+_145241454 | 0.01 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr5_+_117378510 | 0.01 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr10_-_130020042 | 0.01 |
ENSMUST00000216661.2
ENSMUST00000203720.3 |
Olfr826
|
olfactory receptor 826 |
| chr4_+_146586445 | 0.01 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr1_+_88234454 | 0.01 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr14_+_53994813 | 0.01 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr11_+_62737887 | 0.01 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr14_+_53599724 | 0.01 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr5_+_150679760 | 0.01 |
ENSMUST00000110486.2
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr10_+_129493563 | 0.01 |
ENSMUST00000217094.2
|
Olfr800
|
olfactory receptor 800 |
| chr2_+_87686206 | 0.01 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr17_+_38104420 | 0.01 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr7_-_115459082 | 0.01 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr6_-_70116066 | 0.01 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chrX_-_142716085 | 0.01 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr9_+_96140781 | 0.01 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr2_-_87570322 | 0.01 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr13_+_23343482 | 0.01 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chrX_+_9751861 | 0.01 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_66537080 | 0.01 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr4_-_14621497 | 0.01 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_-_146993984 | 0.01 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr4_+_109092610 | 0.01 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr5_-_142515792 | 0.01 |
ENSMUST00000099400.3
|
Papolb
|
poly (A) polymerase beta (testis specific) |
| chr15_-_101422054 | 0.01 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr9_-_15212745 | 0.01 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr2_-_86372824 | 0.01 |
ENSMUST00000216480.2
ENSMUST00000111582.3 |
Olfr1079
|
olfactory receptor 1079 |
| chr1_+_153541412 | 0.01 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr15_-_98507913 | 0.01 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr11_+_67128843 | 0.01 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr7_+_107992374 | 0.01 |
ENSMUST00000215215.2
|
Olfr495
|
olfactory receptor 495 |
| chr3_+_59989282 | 0.01 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr3_+_122213420 | 0.01 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr9_+_38738911 | 0.01 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr2_-_88559941 | 0.01 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr17_-_37404764 | 0.01 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr14_+_51648458 | 0.01 |
ENSMUST00000022438.12
|
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chrX_+_162923474 | 0.01 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr14_-_109151590 | 0.01 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr17_-_14914484 | 0.01 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr16_-_29363671 | 0.01 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr1_+_153541339 | 0.01 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_+_104770653 | 0.01 |
ENSMUST00000106803.9
ENSMUST00000106804.2 |
Fyb2
|
FYN binding protein 2 |
| chr7_-_86046700 | 0.01 |
ENSMUST00000215733.2
|
Olfr303
|
olfactory receptor 303 |
| chr3_-_67422821 | 0.01 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr17_+_37977879 | 0.01 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr11_+_59197746 | 0.01 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr3_-_144514386 | 0.01 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr2_+_111607774 | 0.01 |
ENSMUST00000214708.2
ENSMUST00000215244.2 |
Olfr1302
|
olfactory receptor 1302 |
| chr19_+_13757912 | 0.00 |
ENSMUST00000057390.4
ENSMUST00000219412.2 |
Olfr1496
|
olfactory receptor 1496 |
| chr1_-_166509404 | 0.00 |
ENSMUST00000148677.2
ENSMUST00000027843.11 |
Fmo9
|
flavin containing monooxygenase 9 |
| chr11_+_73489420 | 0.00 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr14_-_48902555 | 0.00 |
ENSMUST00000118578.9
|
Otx2
|
orthodenticle homeobox 2 |
| chr9_+_96140750 | 0.00 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr15_+_98468885 | 0.00 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr7_+_21531776 | 0.00 |
ENSMUST00000170931.2
|
Vmn1r135
|
vomeronasal 1 receptor 135 |
| chr16_-_16494269 | 0.00 |
ENSMUST00000206365.3
|
Olfr19
|
olfactory receptor 19 |
| chr11_-_99979052 | 0.00 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr10_+_129601351 | 0.00 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808 |
| chr2_+_85745537 | 0.00 |
ENSMUST00000213474.3
|
Olfr1025-ps1
|
olfactory receptor 1025, pseudogene 1 |
| chr17_-_68257900 | 0.00 |
ENSMUST00000164647.8
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr10_+_36382810 | 0.00 |
ENSMUST00000167191.8
ENSMUST00000058738.11 |
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr16_-_19443851 | 0.00 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr2_-_76700830 | 0.00 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr2_+_86612823 | 0.00 |
ENSMUST00000213354.2
ENSMUST00000219561.2 |
Olfr1093
|
olfactory receptor 1093 |
| chr1_+_21310803 | 0.00 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr3_+_93184854 | 0.00 |
ENSMUST00000180308.3
|
Flg
|
filaggrin |
| chr2_-_86488800 | 0.00 |
ENSMUST00000099879.2
|
Olfr1085
|
olfactory receptor 1085 |
| chr19_-_39875192 | 0.00 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr2_-_88854443 | 0.00 |
ENSMUST00000099799.5
|
Olfr1217
|
olfactory receptor 1217 |
| chr18_+_57601541 | 0.00 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr7_-_107888141 | 0.00 |
ENSMUST00000211345.3
|
Olfr490
|
olfactory receptor 490 |
| chr3_+_132335575 | 0.00 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr7_+_102526329 | 0.00 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr7_-_119058489 | 0.00 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr12_-_118930130 | 0.00 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr11_+_62737936 | 0.00 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr3_+_132335704 | 0.00 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr5_+_67126304 | 0.00 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr4_+_88768324 | 0.00 |
ENSMUST00000094972.2
|
Ifna1
|
interferon alpha 1 |
| chr17_+_69746321 | 0.00 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chr16_+_14407073 | 0.00 |
ENSMUST00000100165.4
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr19_-_39801188 | 0.00 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr15_+_16778187 | 0.00 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr14_+_96118660 | 0.00 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr4_+_145397238 | 0.00 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr17_-_37615204 | 0.00 |
ENSMUST00000214376.2
|
Olfr101
|
olfactory receptor 101 |
| chr4_+_145595364 | 0.00 |
ENSMUST00000123460.2
|
Zfp986
|
zinc finger protein 986 |
| chr10_+_129357661 | 0.00 |
ENSMUST00000217228.2
|
Olfr791
|
olfactory receptor 791 |
| chr2_+_87405430 | 0.00 |
ENSMUST00000081034.2
|
Olfr1129
|
olfactory receptor 1129 |
| chrX_+_151320497 | 0.00 |
ENSMUST00000238455.2
|
Cldn34a
|
claudin 34A |
| chr3_+_135053762 | 0.00 |
ENSMUST00000159658.8
ENSMUST00000078568.12 |
Slc9b1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr11_-_49004584 | 0.00 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr7_-_37472290 | 0.00 |
ENSMUST00000176205.8
|
Zfp536
|
zinc finger protein 536 |
| chr7_+_22754965 | 0.00 |
ENSMUST00000164537.2
|
Vmn1r163
|
vomeronasal 1 receptor 163 |
| chr9_+_18320390 | 0.00 |
ENSMUST00000098973.3
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr2_+_81883566 | 0.00 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr6_-_115569504 | 0.00 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr11_-_83959999 | 0.00 |
ENSMUST00000138208.2
|
Dusp14
|
dual specificity phosphatase 14 |
| chr5_-_74863514 | 0.00 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr11_+_76792977 | 0.00 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chrX_-_21041892 | 0.00 |
ENSMUST00000096511.5
|
Gm21876
|
predicted gene, 21876 |
| chr6_-_50631418 | 0.00 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr7_+_107166925 | 0.00 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr1_-_158183894 | 0.00 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr12_+_29584560 | 0.00 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr18_-_42132106 | 0.00 |
ENSMUST00000097591.5
|
Grxcr2
|
glutaredoxin, cysteine rich 2 |
| chr6_-_128252540 | 0.00 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr7_+_20606181 | 0.00 |
ENSMUST00000164288.2
|
Vmn1r116
|
vomeronasal 1 receptor 116 |
| chr7_-_5325456 | 0.00 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr7_+_43077088 | 0.00 |
ENSMUST00000239023.2
|
Gm38999
|
predicted gene, 38999 |
| chr18_-_39000056 | 0.00 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr5_-_70999547 | 0.00 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr11_+_58311921 | 0.00 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr7_+_86312872 | 0.00 |
ENSMUST00000081474.2
|
Olfr293
|
olfactory receptor 293 |
| chr11_+_70410445 | 0.00 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr1_-_37955569 | 0.00 |
ENSMUST00000078307.7
|
Lyg2
|
lysozyme G-like 2 |
| chr5_+_143166759 | 0.00 |
ENSMUST00000031574.10
|
Spdye4b
|
speedy/RINGO cell cycle regulator family, member E4B |
| chr5_-_104225458 | 0.00 |
ENSMUST00000198485.5
ENSMUST00000164471.8 ENSMUST00000178967.2 |
Gm17660
|
predicted gene, 17660 |
| chr18_+_20569226 | 0.00 |
ENSMUST00000019426.5
|
Dsg4
|
desmoglein 4 |
| chr15_-_103473481 | 0.00 |
ENSMUST00000228060.2
ENSMUST00000228895.2 ENSMUST00000023134.5 |
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr4_+_146033882 | 0.00 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:1904793 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |