Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
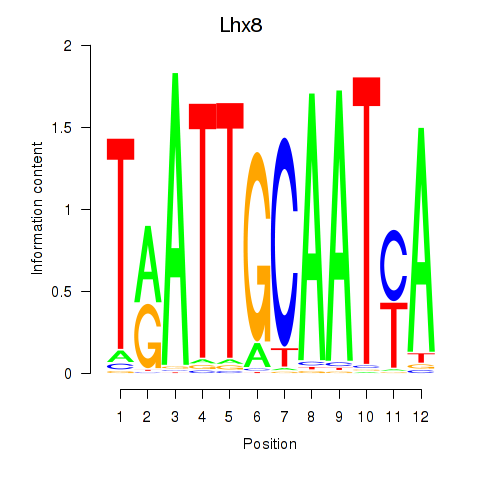
Results for Lhx8
Z-value: 0.91
Transcription factors associated with Lhx8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx8
|
ENSMUSG00000096225.8 | LIM homeobox protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx8 | mm39_v1_chr3_-_154036180_154036296 | 0.82 | 8.8e-02 | Click! |
Activity profile of Lhx8 motif
Sorted Z-values of Lhx8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_44258112 | 1.64 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr16_-_48592372 | 0.77 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr7_+_103379581 | 0.70 |
ENSMUST00000098193.4
|
Olfr628
|
olfactory receptor 628 |
| chrX_-_156198282 | 0.63 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr7_+_140261864 | 0.61 |
ENSMUST00000214637.3
|
Olfr45
|
olfactory receptor 45 |
| chr14_+_54862762 | 0.61 |
ENSMUST00000097177.5
|
Psmb11
|
proteasome (prosome, macropain) subunit, beta type, 11 |
| chr14_+_51648277 | 0.59 |
ENSMUST00000159674.4
ENSMUST00000163019.9 ENSMUST00000233394.2 ENSMUST00000232951.2 ENSMUST00000232797.2 ENSMUST00000233766.2 ENSMUST00000233848.2 ENSMUST00000233280.2 ENSMUST00000233920.2 ENSMUST00000233126.2 ENSMUST00000233651.2 |
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr9_-_40004854 | 0.58 |
ENSMUST00000050996.6
ENSMUST00000213087.4 |
Olfr983
|
olfactory receptor 983 |
| chr4_-_155729865 | 0.57 |
ENSMUST00000115821.3
|
Gm10563
|
predicted gene 10563 |
| chr9_+_38164070 | 0.54 |
ENSMUST00000213129.2
|
Olfr143
|
olfactory receptor 143 |
| chr7_-_12002196 | 0.51 |
ENSMUST00000227973.2
ENSMUST00000228764.2 ENSMUST00000228482.2 ENSMUST00000227080.2 |
Vmn1r81
|
vomeronasal 1 receptor 81 |
| chr8_+_105558204 | 0.48 |
ENSMUST00000059449.7
|
Ces2b
|
carboxyesterase 2B |
| chr5_-_62923463 | 0.47 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr1_+_92545510 | 0.46 |
ENSMUST00000213247.2
|
Olfr12
|
olfactory receptor 12 |
| chr9_+_38202403 | 0.43 |
ENSMUST00000216276.2
ENSMUST00000215219.2 |
Olfr896-ps1
|
olfactory receptor 896, pseudogene 1 |
| chr6_+_15185202 | 0.40 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr7_-_106709576 | 0.40 |
ENSMUST00000215713.2
|
Olfr715b
|
olfactory receptor 715B |
| chr2_-_87838612 | 0.39 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr19_-_12302465 | 0.38 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chr16_+_24540599 | 0.37 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr19_+_13208692 | 0.36 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr2_+_36263531 | 0.35 |
ENSMUST00000072114.4
ENSMUST00000217511.2 |
Olfr338
|
olfactory receptor 338 |
| chr10_-_4382311 | 0.35 |
ENSMUST00000126102.8
ENSMUST00000131853.2 ENSMUST00000042251.11 |
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr2_+_85545763 | 0.35 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr10_-_129577771 | 0.34 |
ENSMUST00000215142.3
ENSMUST00000213239.2 |
Olfr806
|
olfactory receptor 806 |
| chr17_-_47143214 | 0.33 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr4_-_95965767 | 0.32 |
ENSMUST00000030305.13
ENSMUST00000107078.9 |
Cyp2j13
|
cytochrome P450, family 2, subfamily j, polypeptide 13 |
| chr11_-_73382303 | 0.31 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr10_-_4382283 | 0.31 |
ENSMUST00000155172.8
|
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr2_+_89842475 | 0.31 |
ENSMUST00000214382.2
ENSMUST00000217065.3 |
Olfr1263
|
olfactory receptor 1263 |
| chr17_+_33848054 | 0.29 |
ENSMUST00000166627.8
ENSMUST00000073570.12 ENSMUST00000170225.3 |
Zfp414
|
zinc finger protein 414 |
| chr2_-_111880531 | 0.29 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr2_-_36782948 | 0.29 |
ENSMUST00000217215.3
|
Olfr353
|
olfactory receptor 353 |
| chr7_+_43701714 | 0.28 |
ENSMUST00000079859.7
|
Klk1b27
|
kallikrein 1-related peptidase b27 |
| chr11_+_110858842 | 0.26 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_144466602 | 0.25 |
ENSMUST00000059091.6
|
Clca3a1
|
chloride channel accessory 3A1 |
| chr2_-_104680104 | 0.25 |
ENSMUST00000028593.11
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr7_+_12711760 | 0.24 |
ENSMUST00000108535.8
ENSMUST00000108537.8 ENSMUST00000045810.15 |
Zfp446
|
zinc finger protein 446 |
| chr14_+_24540745 | 0.23 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr11_-_109886601 | 0.23 |
ENSMUST00000020948.15
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr7_-_106563137 | 0.23 |
ENSMUST00000213552.2
ENSMUST00000040983.6 ENSMUST00000213651.2 |
Olfr6
|
olfactory receptor 6 |
| chrY_-_2663658 | 0.22 |
ENSMUST00000091178.2
|
Sry
|
sex determining region of Chr Y |
| chr1_+_163875783 | 0.22 |
ENSMUST00000027874.6
|
Sele
|
selectin, endothelial cell |
| chr12_-_56583582 | 0.22 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr14_-_30213408 | 0.21 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr17_-_24851581 | 0.20 |
ENSMUST00000226398.2
ENSMUST00000226284.2 ENSMUST00000097373.2 ENSMUST00000228412.2 |
Tsc2
|
TSC complex subunit 2 |
| chr2_-_120867232 | 0.19 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr3_-_57599956 | 0.19 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chr12_-_113379925 | 0.19 |
ENSMUST00000194162.6
ENSMUST00000192250.2 |
Ighd
|
immunoglobulin heavy constant delta |
| chr7_-_28071658 | 0.18 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr17_-_24851549 | 0.17 |
ENSMUST00000227607.2
ENSMUST00000227509.2 ENSMUST00000227745.2 |
Tsc2
|
TSC complex subunit 2 |
| chr7_-_104677667 | 0.17 |
ENSMUST00000215899.2
ENSMUST00000214318.3 |
Olfr675
|
olfactory receptor 675 |
| chr15_-_98118858 | 0.17 |
ENSMUST00000142443.8
ENSMUST00000170618.8 |
Gm44579
Olfr287
|
predicted gene 44579 olfactory receptor 287 |
| chr12_+_112940086 | 0.16 |
ENSMUST00000165079.8
ENSMUST00000221500.2 ENSMUST00000221104.2 ENSMUST00000002880.7 ENSMUST00000222209.2 |
Btbd6
|
BTB (POZ) domain containing 6 |
| chr5_-_109207435 | 0.16 |
ENSMUST00000164875.3
|
Vmn2r11
|
vomeronasal 2, receptor 11 |
| chr2_+_73142945 | 0.15 |
ENSMUST00000090811.11
ENSMUST00000112050.2 |
Scrn3
|
secernin 3 |
| chr7_-_28071919 | 0.15 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr16_+_35361635 | 0.15 |
ENSMUST00000120756.8
|
Sema5b
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr8_+_72973560 | 0.14 |
ENSMUST00000003123.10
|
Fam32a
|
family with sequence similarity 32, member A |
| chr12_+_84034628 | 0.14 |
ENSMUST00000021649.8
|
Acot2
|
acyl-CoA thioesterase 2 |
| chr14_-_36628263 | 0.13 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr7_-_104628324 | 0.13 |
ENSMUST00000217091.2
ENSMUST00000210963.3 |
Olfr671
|
olfactory receptor 671 |
| chr4_+_19818718 | 0.13 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr13_-_21744063 | 0.12 |
ENSMUST00000217519.2
|
Olfr1535
|
olfactory receptor 1535 |
| chr1_+_33947250 | 0.12 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr19_-_17333972 | 0.12 |
ENSMUST00000174236.8
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr3_+_116653113 | 0.11 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr5_-_31337193 | 0.11 |
ENSMUST00000201428.2
ENSMUST00000201468.4 ENSMUST00000101411.6 |
Gtf3c2
Gm29609
|
general transcription factor IIIC, polypeptide 2, beta predicted gene 29609 |
| chr7_-_28072022 | 0.10 |
ENSMUST00000144700.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr9_-_78285942 | 0.10 |
ENSMUST00000034900.8
|
Ooep
|
oocyte expressed protein |
| chr10_+_129184719 | 0.09 |
ENSMUST00000213970.2
|
Olfr782
|
olfactory receptor 782 |
| chr6_-_136150491 | 0.09 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr5_+_125080504 | 0.09 |
ENSMUST00000197746.2
|
Rflna
|
refilin A |
| chr14_+_73380485 | 0.09 |
ENSMUST00000170677.8
ENSMUST00000167401.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_+_172994841 | 0.09 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr14_+_73380163 | 0.09 |
ENSMUST00000170368.8
ENSMUST00000171767.8 ENSMUST00000163533.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr3_+_107198528 | 0.09 |
ENSMUST00000029502.14
|
Slc16a4
|
solute carrier family 16 (monocarboxylic acid transporters), member 4 |
| chr10_+_129219952 | 0.09 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr4_-_129015682 | 0.09 |
ENSMUST00000139450.8
ENSMUST00000125931.9 ENSMUST00000116444.10 |
Hpca
|
hippocalcin |
| chr9_+_118931532 | 0.08 |
ENSMUST00000165231.8
ENSMUST00000140326.8 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr12_-_114621406 | 0.07 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr1_+_21310821 | 0.07 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr5_+_124068728 | 0.07 |
ENSMUST00000094320.10
ENSMUST00000165148.4 |
Ccdc62
|
coiled-coil domain containing 62 |
| chr5_-_139115417 | 0.07 |
ENSMUST00000026973.14
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr19_-_34579356 | 0.07 |
ENSMUST00000168254.3
|
Ifit1bl1
|
interferon induced protein with tetratricpeptide repeats 1B like 1 |
| chr7_-_3218968 | 0.07 |
ENSMUST00000171749.3
|
AU018091
|
expressed sequence AU018091 |
| chr3_-_72964276 | 0.07 |
ENSMUST00000192477.2
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr14_+_73380577 | 0.06 |
ENSMUST00000165567.8
ENSMUST00000022702.13 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr1_-_74627264 | 0.06 |
ENSMUST00000066986.13
|
Zfp142
|
zinc finger protein 142 |
| chr15_-_99717956 | 0.06 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr2_-_88562710 | 0.06 |
ENSMUST00000213118.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr12_-_115766700 | 0.06 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr14_+_73380539 | 0.06 |
ENSMUST00000169513.8
ENSMUST00000165727.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr17_-_90395771 | 0.06 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr2_+_112114906 | 0.05 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_+_19093665 | 0.05 |
ENSMUST00000140836.8
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr3_+_130411097 | 0.05 |
ENSMUST00000166187.8
ENSMUST00000072271.13 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr12_-_113928438 | 0.04 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr6_+_67701864 | 0.04 |
ENSMUST00000103304.3
|
Igkv1-133
|
immunoglobulin kappa variable 1-133 |
| chr7_-_19093383 | 0.03 |
ENSMUST00000047036.10
|
Cd3eap
|
CD3E antigen, epsilon polypeptide associated protein |
| chr18_+_68470616 | 0.03 |
ENSMUST00000172148.4
ENSMUST00000239469.2 |
Mc5r
|
melanocortin 5 receptor |
| chr7_-_59654849 | 0.03 |
ENSMUST00000059305.17
|
Snrpn
|
small nuclear ribonucleoprotein N |
| chr13_-_62706325 | 0.03 |
ENSMUST00000076195.7
ENSMUST00000101547.12 ENSMUST00000222852.2 ENSMUST00000220648.2 ENSMUST00000222429.2 ENSMUST00000222746.2 |
Zfp935
Zfp934
|
zinc finger protein 935 zinc finger protein 934 |
| chr1_+_161796748 | 0.03 |
ENSMUST00000111594.3
ENSMUST00000028021.7 ENSMUST00000193784.2 ENSMUST00000161826.2 |
Pigc
Gm38304
|
phosphatidylinositol glycan anchor biosynthesis, class C predicted gene, 38304 |
| chr6_+_67586695 | 0.02 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr13_+_23431304 | 0.02 |
ENSMUST00000235331.2
ENSMUST00000236495.2 ENSMUST00000238002.2 |
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr6_+_15184630 | 0.02 |
ENSMUST00000115470.3
|
Foxp2
|
forkhead box P2 |
| chr5_-_123620632 | 0.02 |
ENSMUST00000198901.2
|
Il31
|
interleukin 31 |
| chr13_+_65660492 | 0.02 |
ENSMUST00000081471.3
|
Gm10139
|
predicted gene 10139 |
| chr13_-_62614608 | 0.01 |
ENSMUST00000223247.2
ENSMUST00000221747.2 |
Gm49359
|
predicted gene, 49359 |
| chr13_-_66375387 | 0.01 |
ENSMUST00000167981.3
|
Gm10772
|
predicted gene 10772 |
| chr16_-_58647137 | 0.01 |
ENSMUST00000215069.2
ENSMUST00000079955.6 |
Olfr175
|
olfactory receptor 175 |
| chr14_+_118504372 | 0.01 |
ENSMUST00000171107.3
|
Gm9376
|
predicted gene 9376 |
| chr15_+_54609011 | 0.01 |
ENSMUST00000050027.9
|
Ccn3
|
cellular communication network factor 3 |
| chr1_+_180878797 | 0.01 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr5_+_137551774 | 0.00 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr11_-_73290321 | 0.00 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr19_-_11313471 | 0.00 |
ENSMUST00000056035.9
ENSMUST00000067532.11 |
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr6_-_130258891 | 0.00 |
ENSMUST00000112020.2
|
Klra10
|
killer cell lectin-like receptor subfamily A, member 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |