Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
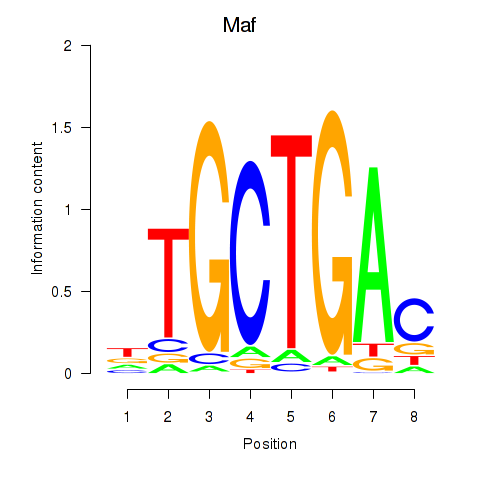
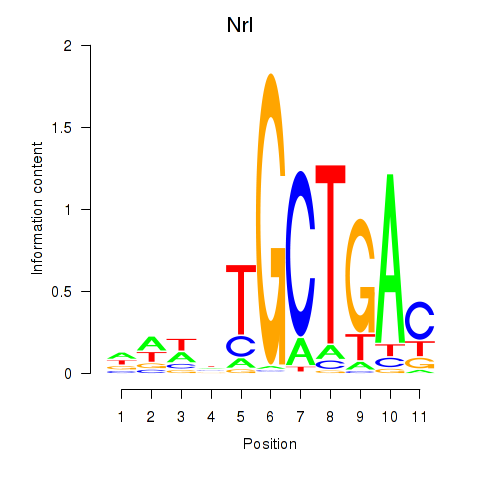
Results for Maf_Nrl
Z-value: 0.61
Transcription factors associated with Maf_Nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maf
|
ENSMUSG00000055435.7 | avian musculoaponeurotic fibrosarcoma oncogene homolog |
|
Nrl
|
ENSMUSG00000040632.17 | neural retina leucine zipper gene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nrl | mm39_v1_chr14_-_55761648_55761680 | -0.27 | 6.7e-01 | Click! |
| Maf | mm39_v1_chr8_-_116434517_116434533 | 0.11 | 8.6e-01 | Click! |
Activity profile of Maf_Nrl motif
Sorted Z-values of Maf_Nrl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_34124078 | 0.64 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr1_+_171386752 | 0.37 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr18_-_61840654 | 0.32 |
ENSMUST00000025472.7
|
Pcyox1l
|
prenylcysteine oxidase 1 like |
| chr14_+_54491637 | 0.24 |
ENSMUST00000180359.8
ENSMUST00000199338.2 |
Abhd4
|
abhydrolase domain containing 4 |
| chr6_-_124441731 | 0.23 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr9_-_110453427 | 0.23 |
ENSMUST00000196876.2
ENSMUST00000035069.14 |
Nradd
|
neurotrophin receptor associated death domain |
| chr2_+_120307390 | 0.21 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr11_+_3280401 | 0.16 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr10_-_14593935 | 0.15 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr12_+_80691275 | 0.15 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr17_-_84495364 | 0.14 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr5_-_77555881 | 0.14 |
ENSMUST00000163898.6
ENSMUST00000046746.10 |
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chr3_-_146476331 | 0.13 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr1_+_127234441 | 0.12 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr11_+_3280771 | 0.12 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr8_-_23680954 | 0.12 |
ENSMUST00000209507.2
|
Gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr6_-_83030759 | 0.11 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr16_-_18052937 | 0.11 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr6_+_41515152 | 0.11 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr14_-_30909082 | 0.11 |
ENSMUST00000170253.8
|
Nisch
|
nischarin |
| chr13_-_97897139 | 0.11 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr3_+_95801325 | 0.11 |
ENSMUST00000197081.2
ENSMUST00000056710.10 |
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr2_+_101455022 | 0.11 |
ENSMUST00000044031.4
|
Rag2
|
recombination activating gene 2 |
| chr11_+_97732108 | 0.10 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr10_+_128247598 | 0.10 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr18_+_69633741 | 0.10 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr11_+_120123727 | 0.10 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr6_+_68098030 | 0.10 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr11_+_76836330 | 0.10 |
ENSMUST00000021197.10
|
Blmh
|
bleomycin hydrolase |
| chr7_+_18962252 | 0.10 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chrX_+_149377416 | 0.10 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr16_-_56987670 | 0.09 |
ENSMUST00000023432.10
|
Nit2
|
nitrilase family, member 2 |
| chr11_+_101215889 | 0.09 |
ENSMUST00000041095.14
ENSMUST00000107264.2 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr11_-_97040858 | 0.09 |
ENSMUST00000118375.8
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr7_+_127347308 | 0.09 |
ENSMUST00000188580.3
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr12_-_113324852 | 0.09 |
ENSMUST00000223179.2
ENSMUST00000103423.3 |
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr17_+_87270707 | 0.09 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr17_+_6320731 | 0.09 |
ENSMUST00000088940.6
|
Tmem181a
|
transmembrane protein 181A |
| chr6_+_129510331 | 0.08 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_+_127347339 | 0.08 |
ENSMUST00000206893.2
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr16_-_45975440 | 0.08 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr6_+_68233361 | 0.08 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr4_+_154096188 | 0.08 |
ENSMUST00000169622.8
ENSMUST00000030894.15 |
Lrrc47
|
leucine rich repeat containing 47 |
| chr19_+_60744385 | 0.07 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr9_+_44309727 | 0.07 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr7_-_92319126 | 0.07 |
ENSMUST00000119954.9
|
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr2_-_32584132 | 0.07 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr4_-_135300934 | 0.07 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr9_+_7272514 | 0.07 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr1_-_97689263 | 0.07 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_-_69771797 | 0.07 |
ENSMUST00000238978.2
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chr2_-_26028814 | 0.07 |
ENSMUST00000163836.2
|
Tmem250-ps
|
transmembrane protein 250, pseudogene |
| chr10_+_41763423 | 0.07 |
ENSMUST00000041438.7
|
Sesn1
|
sestrin 1 |
| chr5_-_138153956 | 0.07 |
ENSMUST00000132318.2
ENSMUST00000049393.15 |
Zfp113
|
zinc finger protein 113 |
| chr5_+_110801310 | 0.07 |
ENSMUST00000031478.6
|
Ddx51
|
DEAD box helicase 51 |
| chr2_+_51928017 | 0.07 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr1_+_85538554 | 0.07 |
ENSMUST00000162925.2
|
Sp140
|
Sp140 nuclear body protein |
| chr16_+_87251852 | 0.07 |
ENSMUST00000119504.8
ENSMUST00000131356.8 |
Usp16
|
ubiquitin specific peptidase 16 |
| chr13_+_84370405 | 0.07 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr17_+_25992761 | 0.07 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr12_+_35097179 | 0.06 |
ENSMUST00000163677.3
ENSMUST00000048519.17 |
Snx13
|
sorting nexin 13 |
| chr19_+_56414114 | 0.06 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr19_+_38252984 | 0.06 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chrX_+_41241049 | 0.06 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr5_+_110692162 | 0.06 |
ENSMUST00000040001.14
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr6_+_145091196 | 0.06 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr17_-_48189815 | 0.06 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4 |
| chr18_+_21134928 | 0.06 |
ENSMUST00000234536.2
ENSMUST00000052396.7 |
Rnf138
|
ring finger protein 138 |
| chr1_+_127657142 | 0.06 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr10_+_83196070 | 0.06 |
ENSMUST00000020488.9
|
D10Wsu102e
|
DNA segment, Chr 10, Wayne State University 102, expressed |
| chr13_+_83652352 | 0.06 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_-_12745109 | 0.06 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr6_-_68681962 | 0.06 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr7_+_100970435 | 0.06 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr12_+_108572015 | 0.06 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr5_-_125371162 | 0.06 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr11_+_77576981 | 0.05 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr16_+_17149235 | 0.05 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr16_+_8647959 | 0.05 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr11_+_106107752 | 0.05 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr18_-_35760260 | 0.05 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr14_+_32507920 | 0.05 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr5_-_25113358 | 0.05 |
ENSMUST00000114975.8
ENSMUST00000150135.8 |
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_+_128247492 | 0.05 |
ENSMUST00000171342.3
|
Rnf41
|
ring finger protein 41 |
| chr12_+_80690985 | 0.05 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr5_-_112400375 | 0.05 |
ENSMUST00000112385.8
|
Cryba4
|
crystallin, beta A4 |
| chr10_+_87694924 | 0.05 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr6_-_83031358 | 0.05 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr18_-_36903237 | 0.05 |
ENSMUST00000235494.2
|
Hars
|
histidyl-tRNA synthetase |
| chr17_-_25394445 | 0.05 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr11_-_107228382 | 0.05 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr6_-_76474767 | 0.05 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr8_+_106587268 | 0.05 |
ENSMUST00000212610.2
ENSMUST00000212484.2 ENSMUST00000212200.2 |
Nutf2
|
nuclear transport factor 2 |
| chr4_-_129271909 | 0.05 |
ENSMUST00000030610.3
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr18_-_10610048 | 0.05 |
ENSMUST00000115864.8
ENSMUST00000145320.2 ENSMUST00000097670.10 |
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr12_+_30634425 | 0.05 |
ENSMUST00000057151.10
|
Tmem18
|
transmembrane protein 18 |
| chr12_-_85871281 | 0.05 |
ENSMUST00000021676.12
ENSMUST00000142331.2 |
Erg28
|
ergosterol biosynthesis 28 |
| chr3_-_95135946 | 0.05 |
ENSMUST00000065482.6
|
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr18_+_21135079 | 0.05 |
ENSMUST00000234545.2
|
Rnf138
|
ring finger protein 138 |
| chr4_-_43578823 | 0.05 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr17_-_48716756 | 0.05 |
ENSMUST00000160319.8
ENSMUST00000159535.2 ENSMUST00000078800.13 ENSMUST00000046719.14 ENSMUST00000162460.8 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr10_+_69048914 | 0.05 |
ENSMUST00000163497.8
ENSMUST00000164212.8 ENSMUST00000067908.14 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr7_+_30159137 | 0.05 |
ENSMUST00000006825.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr15_+_82012114 | 0.05 |
ENSMUST00000089174.6
|
Ccdc134
|
coiled-coil domain containing 134 |
| chr7_+_140438468 | 0.05 |
ENSMUST00000209766.2
|
Ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr14_+_32043944 | 0.04 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr2_+_96148418 | 0.04 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr10_+_115979787 | 0.04 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_-_93146825 | 0.04 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr11_-_78950641 | 0.04 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr12_+_108860192 | 0.04 |
ENSMUST00000220667.2
ENSMUST00000167816.8 ENSMUST00000047115.9 |
Wdr25
|
WD repeat domain 25 |
| chr5_+_67765216 | 0.04 |
ENSMUST00000087241.7
|
Shisa3
|
shisa family member 3 |
| chr11_+_76836545 | 0.04 |
ENSMUST00000125145.8
|
Blmh
|
bleomycin hydrolase |
| chr13_+_83652280 | 0.04 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_-_127147609 | 0.04 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr9_-_39486831 | 0.04 |
ENSMUST00000216298.2
ENSMUST00000215194.2 |
Olfr959
|
olfactory receptor 959 |
| chr18_-_35788255 | 0.04 |
ENSMUST00000190196.5
|
Prob1
|
proline rich basic protein 1 |
| chr14_+_67470884 | 0.04 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr6_+_70495224 | 0.04 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr2_+_151385797 | 0.04 |
ENSMUST00000142271.2
|
Fkbp1a
|
FK506 binding protein 1a |
| chr6_+_33226020 | 0.04 |
ENSMUST00000052266.15
ENSMUST00000090381.11 ENSMUST00000115080.2 |
Exoc4
|
exocyst complex component 4 |
| chr1_-_65142521 | 0.04 |
ENSMUST00000061497.9
|
Cryga
|
crystallin, gamma A |
| chr15_-_85387428 | 0.04 |
ENSMUST00000178942.2
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr13_+_52750883 | 0.04 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr7_-_92319096 | 0.04 |
ENSMUST00000208255.2
ENSMUST00000208058.2 |
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chrX_-_93166992 | 0.04 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr5_-_129856267 | 0.04 |
ENSMUST00000201394.4
|
Psph
|
phosphoserine phosphatase |
| chr5_-_92404137 | 0.04 |
ENSMUST00000201130.4
|
Ppef2
|
protein phosphatase, EF hand calcium-binding domain 2 |
| chr6_+_17491231 | 0.03 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr1_-_151965876 | 0.03 |
ENSMUST00000044581.14
|
1700025G04Rik
|
RIKEN cDNA 1700025G04 gene |
| chr19_-_4447080 | 0.03 |
ENSMUST00000075856.11
ENSMUST00000176483.3 ENSMUST00000116571.9 |
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr2_+_58991182 | 0.03 |
ENSMUST00000168631.8
ENSMUST00000102754.11 ENSMUST00000123908.8 |
Pkp4
|
plakophilin 4 |
| chr3_+_96739458 | 0.03 |
ENSMUST00000107070.8
ENSMUST00000107069.8 |
Pdzk1
|
PDZ domain containing 1 |
| chr6_+_70675416 | 0.03 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr12_-_113700190 | 0.03 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr7_-_141456092 | 0.03 |
ENSMUST00000055819.13
ENSMUST00000001950.12 |
Tollip
|
toll interacting protein |
| chr9_+_44966464 | 0.03 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr11_-_109364424 | 0.03 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr7_-_126062272 | 0.03 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr16_-_38115172 | 0.03 |
ENSMUST00000023504.5
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr16_-_62667307 | 0.03 |
ENSMUST00000232561.2
ENSMUST00000089289.6 |
Arl13b
|
ADP-ribosylation factor-like 13B |
| chr7_+_18962301 | 0.03 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr8_-_125624424 | 0.03 |
ENSMUST00000098312.4
|
Exoc8
|
exocyst complex component 8 |
| chr19_-_21630143 | 0.03 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr5_-_31117617 | 0.03 |
ENSMUST00000202567.2
ENSMUST00000074840.12 |
Preb
|
prolactin regulatory element binding |
| chr4_-_126096376 | 0.03 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr17_+_64203017 | 0.03 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr17_+_26882171 | 0.03 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr10_+_79716876 | 0.03 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr6_+_142359099 | 0.03 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr2_+_29533799 | 0.03 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_+_107656810 | 0.02 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr9_+_109881083 | 0.02 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr16_+_21644692 | 0.02 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr10_+_127157784 | 0.02 |
ENSMUST00000219511.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr13_+_83652150 | 0.02 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr9_-_40257586 | 0.02 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr11_-_107361525 | 0.02 |
ENSMUST00000103064.10
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_+_100971034 | 0.02 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr2_+_34661982 | 0.02 |
ENSMUST00000028222.13
ENSMUST00000100171.3 |
Hspa5
|
heat shock protein 5 |
| chr2_+_58991352 | 0.02 |
ENSMUST00000037903.15
|
Pkp4
|
plakophilin 4 |
| chr1_-_9818601 | 0.02 |
ENSMUST00000057438.7
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr13_+_16189041 | 0.02 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr16_+_20367327 | 0.02 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr6_+_88061464 | 0.02 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr15_+_10224052 | 0.02 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr5_+_34731008 | 0.02 |
ENSMUST00000114338.9
|
Add1
|
adducin 1 (alpha) |
| chr15_-_66985760 | 0.02 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr7_+_100970910 | 0.02 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr13_+_42862957 | 0.02 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr12_-_108860042 | 0.02 |
ENSMUST00000109848.10
|
Wars
|
tryptophanyl-tRNA synthetase |
| chr11_+_4959515 | 0.02 |
ENSMUST00000101613.3
|
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr19_+_20579322 | 0.02 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr10_+_56253418 | 0.02 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr18_-_35788240 | 0.02 |
ENSMUST00000097619.2
|
Prob1
|
proline rich basic protein 1 |
| chr18_-_60866186 | 0.02 |
ENSMUST00000237885.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr9_-_38585897 | 0.02 |
ENSMUST00000215461.3
|
Olfr918
|
olfactory receptor 918 |
| chr7_+_107264518 | 0.02 |
ENSMUST00000239294.2
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr10_+_86541675 | 0.02 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr8_-_72175949 | 0.02 |
ENSMUST00000125092.2
|
Fcho1
|
FCH domain only 1 |
| chr16_-_17711950 | 0.02 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr5_-_24628483 | 0.02 |
ENSMUST00000198990.2
|
Cdk5
|
cyclin-dependent kinase 5 |
| chrX_+_41591476 | 0.02 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr6_-_69835868 | 0.02 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr18_+_5593566 | 0.02 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr6_-_40531984 | 0.02 |
ENSMUST00000215009.3
|
Olfr461
|
olfactory receptor 461 |
| chr2_-_30720345 | 0.02 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr4_+_59003121 | 0.02 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr2_+_26481435 | 0.01 |
ENSMUST00000152988.9
ENSMUST00000149789.2 |
Egfl7
|
EGF-like domain 7 |
| chr10_-_24588030 | 0.01 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_-_80255156 | 0.01 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr18_+_36693646 | 0.01 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr7_+_27967222 | 0.01 |
ENSMUST00000059596.8
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr7_+_44866635 | 0.01 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr3_+_45332831 | 0.01 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr11_-_53313950 | 0.01 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr4_+_155924181 | 0.01 |
ENSMUST00000030947.4
|
Mxra8
|
matrix-remodelling associated 8 |
| chr9_+_64024429 | 0.01 |
ENSMUST00000034969.14
|
Lctl
|
lactase-like |
| chr8_+_47192911 | 0.01 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr1_+_180396478 | 0.01 |
ENSMUST00000027777.12
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Maf_Nrl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0002358 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.0 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.0 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |