Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
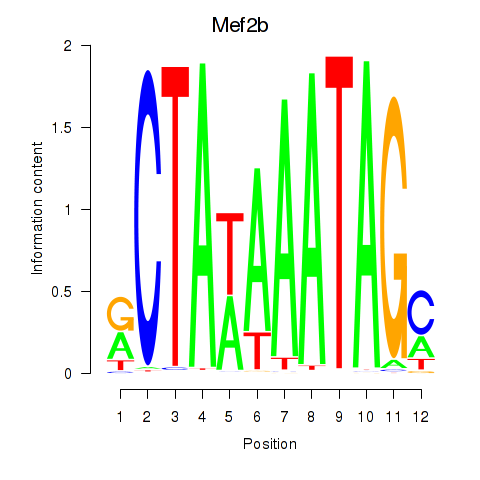
Results for Mef2b
Z-value: 0.43
Transcription factors associated with Mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2b
|
ENSMUSG00000079033.11 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2b | mm39_v1_chr8_+_70605404_70605431 | -0.96 | 1.0e-02 | Click! |
Activity profile of Mef2b motif
Sorted Z-values of Mef2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_100164007 | 0.22 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr17_-_26056056 | 0.22 |
ENSMUST00000183929.8
ENSMUST00000184865.2 ENSMUST00000026831.14 |
Rhbdl1
|
rhomboid like 1 |
| chr2_-_85966272 | 0.21 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr6_-_3399451 | 0.18 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr5_+_115034997 | 0.18 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr7_-_104570689 | 0.14 |
ENSMUST00000217177.2
|
Olfr667
|
olfactory receptor 667 |
| chr8_+_105688344 | 0.13 |
ENSMUST00000043183.8
|
Ces2g
|
carboxylesterase 2G |
| chr1_-_173770010 | 0.12 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr6_-_112924205 | 0.12 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr12_-_34956910 | 0.12 |
ENSMUST00000239321.2
|
Hdac9
|
histone deacetylase 9 |
| chr4_+_156300325 | 0.12 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr17_-_33049169 | 0.12 |
ENSMUST00000201499.2
ENSMUST00000201876.4 ENSMUST00000202759.2 ENSMUST00000202988.4 |
Zfp799
|
zinc finger protein 799 |
| chr6_-_112923715 | 0.11 |
ENSMUST00000113169.9
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr13_-_23553327 | 0.11 |
ENSMUST00000125328.2
ENSMUST00000145451.8 ENSMUST00000050101.9 |
Zfp322a
|
zinc finger protein 322A |
| chr7_-_12552764 | 0.11 |
ENSMUST00000108546.2
ENSMUST00000072222.8 |
Zfp329
|
zinc finger protein 329 |
| chr11_+_73051228 | 0.11 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_+_137849189 | 0.11 |
ENSMUST00000040321.13
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr13_+_5911481 | 0.10 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr1_+_36411312 | 0.10 |
ENSMUST00000179162.7
|
Fer1l5
|
fer-1-like 5 (C. elegans) |
| chr18_-_25887173 | 0.10 |
ENSMUST00000225477.2
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr18_+_24087725 | 0.10 |
ENSMUST00000225682.2
ENSMUST00000060762.6 |
Zfp397
|
zinc finger protein 397 |
| chr17_+_19880590 | 0.10 |
ENSMUST00000171741.3
|
Vmn2r102
|
vomeronasal 2, receptor 102 |
| chr6_-_131224305 | 0.10 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr9_-_14651985 | 0.09 |
ENSMUST00000076946.4
ENSMUST00000115644.10 |
Piwil4
|
piwi-like RNA-mediated gene silencing 4 |
| chr14_-_51134930 | 0.09 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr5_-_129030367 | 0.09 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr5_+_23992689 | 0.09 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr2_+_120294046 | 0.09 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr7_+_81512421 | 0.09 |
ENSMUST00000119543.2
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr1_+_177273226 | 0.09 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_+_9618173 | 0.08 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr8_+_23548541 | 0.08 |
ENSMUST00000173248.8
|
Ank1
|
ankyrin 1, erythroid |
| chr1_-_65158717 | 0.08 |
ENSMUST00000144760.3
|
Gm28845
|
predicted gene 28845 |
| chr7_-_44393654 | 0.08 |
ENSMUST00000149011.9
|
Zfp473
|
zinc finger protein 473 |
| chr13_-_23553139 | 0.08 |
ENSMUST00000152557.8
|
Zfp322a
|
zinc finger protein 322A |
| chr4_-_115911053 | 0.08 |
ENSMUST00000030475.3
|
Nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr4_-_9643636 | 0.08 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr12_+_71095112 | 0.08 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_+_34258411 | 0.08 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr19_-_6002210 | 0.07 |
ENSMUST00000236013.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr19_+_6144449 | 0.07 |
ENSMUST00000235513.2
|
Gm550
|
predicted gene 550 |
| chr3_-_154302679 | 0.07 |
ENSMUST00000052774.8
ENSMUST00000170461.8 ENSMUST00000122976.2 |
Tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr11_+_119283887 | 0.07 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr11_+_88890202 | 0.07 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr6_+_137387718 | 0.07 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr11_-_99045894 | 0.07 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr15_-_89310060 | 0.06 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr2_-_111820618 | 0.06 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr1_+_10109987 | 0.06 |
ENSMUST00000118263.8
ENSMUST00000122156.8 ENSMUST00000119714.8 |
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr18_-_25886908 | 0.06 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr12_-_81426238 | 0.06 |
ENSMUST00000062182.8
|
Gm4787
|
predicted gene 4787 |
| chr18_+_60426444 | 0.06 |
ENSMUST00000171297.2
|
F830016B08Rik
|
RIKEN cDNA F830016B08 gene |
| chr5_+_92719336 | 0.06 |
ENSMUST00000176621.8
ENSMUST00000175974.2 ENSMUST00000131166.9 ENSMUST00000176448.8 ENSMUST00000082382.8 |
Fam47e
|
family with sequence similarity 47, member E |
| chr3_-_86906591 | 0.06 |
ENSMUST00000063869.11
ENSMUST00000029717.4 |
Cd1d1
|
CD1d1 antigen |
| chr16_+_31998588 | 0.06 |
ENSMUST00000231941.2
|
Bex6
|
brain expressed family member 6 |
| chr13_-_74512111 | 0.06 |
ENSMUST00000022064.5
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr15_+_25940859 | 0.05 |
ENSMUST00000226750.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_-_87798643 | 0.05 |
ENSMUST00000099841.4
|
Olfr1157
|
olfactory receptor 1157 |
| chr3_+_96552895 | 0.05 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chrX_-_92875712 | 0.05 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr9_+_123635529 | 0.05 |
ENSMUST00000049810.9
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr7_+_29821340 | 0.05 |
ENSMUST00000098596.11
ENSMUST00000153792.2 |
Zfp382
|
zinc finger protein 382 |
| chr14_-_50564218 | 0.05 |
ENSMUST00000217152.2
|
Olfr734
|
olfactory receptor 734 |
| chr4_+_62581857 | 0.05 |
ENSMUST00000126338.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr2_-_87838612 | 0.05 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr10_+_129306867 | 0.05 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr1_-_173707677 | 0.05 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr7_+_140343652 | 0.05 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr14_+_47886748 | 0.05 |
ENSMUST00000190252.7
ENSMUST00000186466.3 |
Ktn1
|
kinectin 1 |
| chr15_+_73620213 | 0.05 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr6_+_137387729 | 0.05 |
ENSMUST00000203914.2
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr11_+_77692116 | 0.05 |
ENSMUST00000100794.10
ENSMUST00000151373.4 ENSMUST00000130305.9 ENSMUST00000172303.10 ENSMUST00000092884.11 ENSMUST00000164334.8 |
Myo18a
|
myosin XVIIIA |
| chr9_+_123635556 | 0.05 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr15_-_76116245 | 0.05 |
ENSMUST00000167754.8
|
Plec
|
plectin |
| chr2_+_79538124 | 0.04 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr18_+_61096660 | 0.04 |
ENSMUST00000039904.7
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr2_-_57014015 | 0.04 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr8_-_112718888 | 0.04 |
ENSMUST00000034427.12
ENSMUST00000139820.8 |
Adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr19_-_29721012 | 0.04 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr2_-_51039112 | 0.04 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr5_+_104656204 | 0.04 |
ENSMUST00000239420.2
ENSMUST00000239307.3 ENSMUST00000096452.7 |
Thoc2l
|
THO complex subunit 2-like |
| chr19_-_46958001 | 0.04 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr7_-_30523191 | 0.04 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr8_+_3550450 | 0.04 |
ENSMUST00000004683.13
ENSMUST00000160338.2 |
Mcoln1
|
mucolipin 1 |
| chr13_+_38388904 | 0.04 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr2_+_111205867 | 0.04 |
ENSMUST00000062407.6
|
Olfr1284
|
olfactory receptor 1284 |
| chr7_+_27878894 | 0.04 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr5_+_64316771 | 0.04 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr5_-_3943907 | 0.04 |
ENSMUST00000117463.2
ENSMUST00000044746.5 |
Mterf1a
|
mitochondrial transcription termination factor 1a |
| chr4_+_46039202 | 0.04 |
ENSMUST00000156200.8
|
Tmod1
|
tropomodulin 1 |
| chr1_-_13061333 | 0.04 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chrX_-_63320543 | 0.04 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr10_+_58230203 | 0.04 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_-_126718037 | 0.04 |
ENSMUST00000028843.12
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr6_-_124942366 | 0.04 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr17_+_28451674 | 0.03 |
ENSMUST00000002320.16
ENSMUST00000232879.2 ENSMUST00000166744.8 |
Ppard
|
peroxisome proliferator activator receptor delta |
| chr2_-_126718129 | 0.03 |
ENSMUST00000103224.10
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr2_-_26406631 | 0.03 |
ENSMUST00000132820.2
|
Notch1
|
notch 1 |
| chr11_-_79394904 | 0.03 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr8_-_35432783 | 0.03 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr7_+_27879650 | 0.03 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr10_+_111808569 | 0.03 |
ENSMUST00000163048.8
ENSMUST00000174653.2 |
Krr1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr6_+_126916956 | 0.03 |
ENSMUST00000201617.2
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr14_+_55813074 | 0.03 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr9_+_110948492 | 0.03 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr13_-_21826688 | 0.03 |
ENSMUST00000205631.3
|
Olfr11
|
olfactory receptor 11 |
| chr13_+_89687915 | 0.03 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr11_-_86561980 | 0.03 |
ENSMUST00000143991.3
|
Vmp1
|
vacuole membrane protein 1 |
| chr15_+_76211597 | 0.03 |
ENSMUST00000059045.8
|
Exosc4
|
exosome component 4 |
| chr3_-_96721829 | 0.03 |
ENSMUST00000047702.8
|
Cd160
|
CD160 antigen |
| chr1_+_171265103 | 0.03 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr17_-_18498018 | 0.03 |
ENSMUST00000172190.4
ENSMUST00000231815.3 |
Vmn2r94
|
vomeronasal 2, receptor 94 |
| chr7_+_102954855 | 0.03 |
ENSMUST00000214577.2
|
Olfr596
|
olfactory receptor 596 |
| chr11_-_54140462 | 0.03 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr4_-_131695135 | 0.03 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr15_+_101164719 | 0.03 |
ENSMUST00000230814.2
ENSMUST00000023779.8 |
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_-_39775182 | 0.03 |
ENSMUST00000178045.9
ENSMUST00000178563.3 |
Mfsd4b4
|
major facilitator superfamily domain containing 4B4 |
| chr2_-_111880531 | 0.03 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr9_-_37627519 | 0.03 |
ENSMUST00000215727.2
ENSMUST00000211952.3 |
Olfr160
|
olfactory receptor 160 |
| chr9_-_123691077 | 0.03 |
ENSMUST00000182350.3
|
Xcr1
|
chemokine (C motif) receptor 1 |
| chr3_+_40754448 | 0.03 |
ENSMUST00000026858.11
|
Plk4
|
polo like kinase 4 |
| chr1_-_138103021 | 0.03 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_+_75546989 | 0.03 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr7_+_141996067 | 0.03 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr6_-_35516700 | 0.02 |
ENSMUST00000201026.2
ENSMUST00000031866.9 |
Mtpn
|
myotrophin |
| chr15_+_98065039 | 0.02 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr15_-_79967543 | 0.02 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr1_+_160806194 | 0.02 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr9_+_99511549 | 0.02 |
ENSMUST00000131095.8
ENSMUST00000078367.12 ENSMUST00000112885.9 |
Dzip1l
|
DAZ interacting protein 1-like |
| chr9_-_39920878 | 0.02 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr11_-_74228504 | 0.02 |
ENSMUST00000213831.2
|
Olfr410
|
olfactory receptor 410 |
| chr16_+_4825146 | 0.02 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr11_-_69838971 | 0.02 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr1_-_74639723 | 0.02 |
ENSMUST00000127938.8
ENSMUST00000154874.8 |
Rnf25
|
ring finger protein 25 |
| chr9_+_74860133 | 0.02 |
ENSMUST00000215370.2
|
Fam214a
|
family with sequence similarity 214, member A |
| chr7_+_44984681 | 0.02 |
ENSMUST00000085351.7
|
Hrc
|
histidine rich calcium binding protein |
| chr4_+_11485945 | 0.02 |
ENSMUST00000055372.14
ENSMUST00000059914.13 |
Virma
|
vir like m6A methyltransferase associated |
| chr11_-_74228857 | 0.02 |
ENSMUST00000214490.2
|
Olfr410
|
olfactory receptor 410 |
| chr1_-_64160557 | 0.02 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_83672389 | 0.02 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_+_132996237 | 0.02 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr2_+_136555364 | 0.02 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr12_-_56583582 | 0.02 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr11_+_105069591 | 0.02 |
ENSMUST00000106939.9
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr6_+_126916919 | 0.02 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr11_+_70548022 | 0.02 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr15_+_25940781 | 0.02 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr1_-_138102972 | 0.02 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr4_+_11486002 | 0.02 |
ENSMUST00000108307.3
|
Virma
|
vir like m6A methyltransferase associated |
| chr7_-_128020397 | 0.02 |
ENSMUST00000147840.2
|
Rgs10
|
regulator of G-protein signalling 10 |
| chr17_+_29251602 | 0.02 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr17_+_25407352 | 0.02 |
ENSMUST00000039734.12
|
Unkl
|
unkempt family like zinc finger |
| chr18_+_61096597 | 0.02 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr8_-_106670014 | 0.02 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr7_-_25374472 | 0.02 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr9_+_69305211 | 0.02 |
ENSMUST00000117610.8
ENSMUST00000145538.2 ENSMUST00000117246.8 |
Ice2
|
interactor of little elongation complex ELL subunit 2 |
| chrX_+_74139645 | 0.02 |
ENSMUST00000131155.8
ENSMUST00000132000.8 |
Dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr6_+_136530970 | 0.01 |
ENSMUST00000189535.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_+_120614587 | 0.01 |
ENSMUST00000201684.4
ENSMUST00000066540.14 |
Sds
|
serine dehydratase |
| chr17_-_20268598 | 0.01 |
ENSMUST00000168050.3
|
Vmn2r104
|
vomeronasal 2, receptor 104 |
| chr11_+_49327451 | 0.01 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr9_+_123603605 | 0.01 |
ENSMUST00000180093.2
|
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr14_+_26616514 | 0.01 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr17_+_46989695 | 0.01 |
ENSMUST00000233032.2
|
Mea1
|
male enhanced antigen 1 |
| chr14_-_20718337 | 0.01 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr16_+_4825170 | 0.01 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chrX_+_74139460 | 0.01 |
ENSMUST00000033776.15
|
Dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr17_+_29898221 | 0.01 |
ENSMUST00000129864.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr11_-_101315345 | 0.01 |
ENSMUST00000107257.8
ENSMUST00000107259.4 ENSMUST00000107252.9 ENSMUST00000093933.11 |
Gm27029
Ptges3l
|
predicted gene, 27029 prostaglandin E synthase 3 like |
| chr11_+_75546671 | 0.01 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr7_+_31075005 | 0.01 |
ENSMUST00000179481.2
|
Scgb1b3
|
secretoglobin, family 1B, member 3 |
| chr7_-_67022520 | 0.01 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr1_-_160958998 | 0.01 |
ENSMUST00000111611.8
|
Klhl20
|
kelch-like 20 |
| chr5_+_33176160 | 0.01 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr15_+_25940912 | 0.01 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr7_-_127805518 | 0.01 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr12_-_40249489 | 0.01 |
ENSMUST00000220951.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr19_-_11291805 | 0.01 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr11_-_59678462 | 0.01 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr11_-_102120953 | 0.01 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr2_-_33321306 | 0.01 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr7_+_107264518 | 0.01 |
ENSMUST00000239294.2
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_+_172282476 | 0.01 |
ENSMUST00000029005.4
|
Rtf2
|
replication termination factor 2 |
| chr9_-_19799300 | 0.01 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr2_-_181220698 | 0.01 |
ENSMUST00000136875.2
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr16_+_11131676 | 0.01 |
ENSMUST00000023140.6
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr16_-_56533179 | 0.01 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr2_-_111779785 | 0.01 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr3_+_14598927 | 0.01 |
ENSMUST00000163660.8
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr12_-_111638722 | 0.01 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr3_+_14598877 | 0.01 |
ENSMUST00000169079.8
ENSMUST00000091325.10 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr2_+_80122811 | 0.01 |
ENSMUST00000057072.6
|
Prdx6b
|
peroxiredoxin 6B |
| chrX_+_141608694 | 0.01 |
ENSMUST00000112888.2
|
Tmem164
|
transmembrane protein 164 |
| chr14_-_51134906 | 0.01 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr2_-_160169414 | 0.01 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr11_-_102120917 | 0.01 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr4_+_43851565 | 0.01 |
ENSMUST00000107860.3
|
Olfr155
|
olfactory receptor 155 |
| chr15_-_102630589 | 0.01 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr7_+_15863679 | 0.00 |
ENSMUST00000211649.2
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_+_99511876 | 0.00 |
ENSMUST00000112886.9
|
Dzip1l
|
DAZ interacting protein 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:2000547 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0048007 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.0 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |