Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
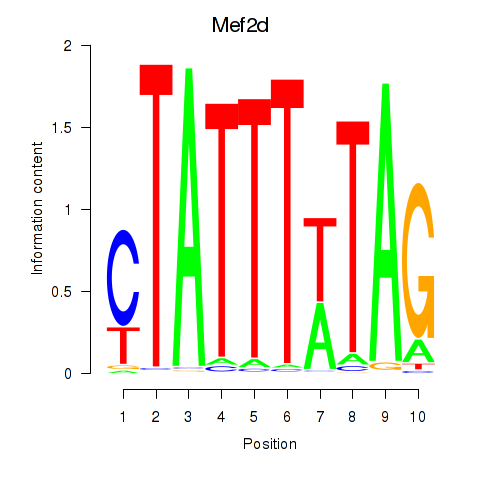
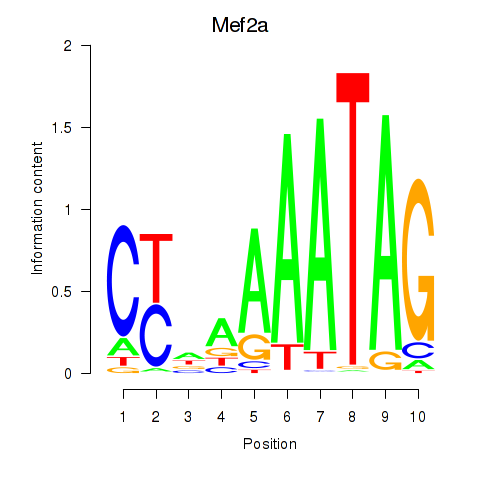
Results for Mef2d_Mef2a
Z-value: 3.05
Transcription factors associated with Mef2d_Mef2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2d
|
ENSMUSG00000001419.18 | myocyte enhancer factor 2D |
|
Mef2a
|
ENSMUSG00000030557.18 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2d | mm39_v1_chr3_+_88049875_88049892 | 0.97 | 5.1e-03 | Click! |
| Mef2a | mm39_v1_chr7_-_67022520_67022609 | 0.79 | 1.1e-01 | Click! |
Activity profile of Mef2d_Mef2a motif
Sorted Z-values of Mef2d_Mef2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_24118139 | 10.28 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr13_-_23946359 | 5.04 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr13_-_23882437 | 4.07 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr5_-_129030367 | 3.28 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr6_+_122803624 | 2.99 |
ENSMUST00000203075.2
|
Foxj2
|
forkhead box J2 |
| chr4_+_62581857 | 2.73 |
ENSMUST00000126338.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr15_+_101164719 | 2.54 |
ENSMUST00000230814.2
ENSMUST00000023779.8 |
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_-_76116245 | 2.26 |
ENSMUST00000167754.8
|
Plec
|
plectin |
| chr3_-_142101339 | 2.17 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr11_-_69289052 | 2.03 |
ENSMUST00000050140.6
|
Tmem88
|
transmembrane protein 88 |
| chr13_+_23868175 | 1.99 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr12_+_71095112 | 1.96 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr5_+_138278777 | 1.92 |
ENSMUST00000048028.15
ENSMUST00000162245.8 ENSMUST00000161691.2 |
Stag3
|
stromal antigen 3 |
| chr12_-_111638722 | 1.85 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr2_-_33261411 | 1.80 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr10_-_127190280 | 1.74 |
ENSMUST00000059718.6
|
Inhbe
|
inhibin beta-E |
| chr2_+_11647610 | 1.74 |
ENSMUST00000028111.6
|
Il2ra
|
interleukin 2 receptor, alpha chain |
| chr5_+_138278502 | 1.73 |
ENSMUST00000160729.8
|
Stag3
|
stromal antigen 3 |
| chr5_+_105667254 | 1.65 |
ENSMUST00000067924.13
ENSMUST00000150981.2 |
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr6_+_116627567 | 1.65 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr2_-_33261498 | 1.62 |
ENSMUST00000113165.8
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr14_-_20718337 | 1.61 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr12_-_34956910 | 1.56 |
ENSMUST00000239321.2
|
Hdac9
|
histone deacetylase 9 |
| chr7_-_127423641 | 1.56 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B |
| chr2_+_3514071 | 1.55 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_-_64161415 | 1.48 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_-_36208265 | 1.47 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr11_+_31822211 | 1.45 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr13_+_89687915 | 1.42 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr9_+_74860133 | 1.41 |
ENSMUST00000215370.2
|
Fam214a
|
family with sequence similarity 214, member A |
| chr9_-_49710058 | 1.38 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_-_107474221 | 1.36 |
ENSMUST00000238519.2
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr16_+_32090286 | 1.35 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr13_-_23867924 | 1.29 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr17_-_66079629 | 1.26 |
ENSMUST00000233258.2
|
Rab31
|
RAB31, member RAS oncogene family |
| chr2_-_111965322 | 1.22 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr19_-_5974825 | 1.22 |
ENSMUST00000055458.6
|
Cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr1_-_89942299 | 1.21 |
ENSMUST00000086882.8
ENSMUST00000097656.10 |
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr3_-_142101418 | 1.15 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr19_-_34856853 | 1.14 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr9_-_49710190 | 1.12 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr14_-_51134930 | 1.11 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr1_+_16758629 | 1.09 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr19_+_38252984 | 1.08 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr13_-_32967937 | 1.07 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr7_+_27878894 | 1.05 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr2_+_71884943 | 1.05 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_-_122752508 | 1.03 |
ENSMUST00000127220.8
ENSMUST00000031426.14 |
Ift81
|
intraflagellar transport 81 |
| chr14_+_47886748 | 1.02 |
ENSMUST00000190252.7
ENSMUST00000186466.3 |
Ktn1
|
kinectin 1 |
| chr12_+_59177552 | 1.01 |
ENSMUST00000175877.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr13_+_5911481 | 0.98 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr11_-_78950698 | 0.94 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr2_+_19662529 | 0.92 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr2_+_48839505 | 0.91 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr2_+_90927053 | 0.88 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_-_64160557 | 0.87 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_+_75742881 | 0.86 |
ENSMUST00000164192.9
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr11_-_83483807 | 0.85 |
ENSMUST00000019071.4
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr13_+_38010879 | 0.84 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr3_+_96484294 | 0.84 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr3_+_90511068 | 0.78 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chr6_+_116627635 | 0.78 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chrX_+_72818003 | 0.78 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr18_-_25887173 | 0.78 |
ENSMUST00000225477.2
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr7_+_27879650 | 0.76 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr5_-_51711237 | 0.75 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr4_-_128699838 | 0.75 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr9_-_101076198 | 0.74 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr15_+_101152078 | 0.74 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr14_-_36641270 | 0.73 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr18_+_61096660 | 0.72 |
ENSMUST00000039904.7
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr9_+_99511549 | 0.72 |
ENSMUST00000131095.8
ENSMUST00000078367.12 ENSMUST00000112885.9 |
Dzip1l
|
DAZ interacting protein 1-like |
| chr9_+_77543776 | 0.72 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr18_+_7905440 | 0.72 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil |
| chr6_+_137387718 | 0.72 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr16_+_14523696 | 0.71 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr15_+_81469538 | 0.70 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chrX_-_48980360 | 0.70 |
ENSMUST00000217355.3
|
Olfr1322
|
olfactory receptor 1322 |
| chr10_-_69048928 | 0.69 |
ENSMUST00000170048.2
|
A930033H14Rik
|
RIKEN cDNA A930033H14 gene |
| chr13_+_102830029 | 0.68 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr12_+_69288606 | 0.68 |
ENSMUST00000063445.13
|
Klhdc1
|
kelch domain containing 1 |
| chr2_+_30485048 | 0.68 |
ENSMUST00000102853.4
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr3_-_103645311 | 0.67 |
ENSMUST00000029440.10
|
Olfml3
|
olfactomedin-like 3 |
| chr6_-_142647944 | 0.67 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_-_119371994 | 0.67 |
ENSMUST00000171024.8
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr17_+_44499451 | 0.67 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr18_+_67597929 | 0.65 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chrX_-_63320543 | 0.65 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr6_+_137387729 | 0.65 |
ENSMUST00000203914.2
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr7_-_67022520 | 0.63 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr11_-_69692542 | 0.62 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr2_-_51039112 | 0.60 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr7_+_29821340 | 0.58 |
ENSMUST00000098596.11
ENSMUST00000153792.2 |
Zfp382
|
zinc finger protein 382 |
| chr13_+_16186410 | 0.58 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr10_-_81037878 | 0.58 |
ENSMUST00000005069.8
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr4_-_41048124 | 0.57 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr7_-_30251680 | 0.57 |
ENSMUST00000215288.2
ENSMUST00000108165.8 ENSMUST00000153594.3 |
Proser3
|
proline and serine rich 3 |
| chr3_-_95049655 | 0.57 |
ENSMUST00000013851.4
|
Tnfaip8l2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr15_-_82796308 | 0.57 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr14_-_20714634 | 0.56 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr12_+_52746158 | 0.56 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr13_+_83721696 | 0.55 |
ENSMUST00000197146.5
ENSMUST00000185052.6 ENSMUST00000195984.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr6_+_43149074 | 0.54 |
ENSMUST00000216179.2
|
Olfr13
|
olfactory receptor 13 |
| chr18_-_25886908 | 0.52 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chrM_+_7006 | 0.52 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr19_-_60779077 | 0.51 |
ENSMUST00000025955.8
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr15_-_80989200 | 0.50 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr19_-_31742427 | 0.50 |
ENSMUST00000065067.14
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr1_-_74788013 | 0.48 |
ENSMUST00000188073.7
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr3_+_137849189 | 0.47 |
ENSMUST00000040321.13
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr2_+_129434834 | 0.46 |
ENSMUST00000103203.8
|
Sirpa
|
signal-regulatory protein alpha |
| chr2_+_129434738 | 0.46 |
ENSMUST00000153491.8
ENSMUST00000161620.8 ENSMUST00000179001.8 |
Sirpa
|
signal-regulatory protein alpha |
| chr19_-_46315543 | 0.46 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr8_+_23548541 | 0.45 |
ENSMUST00000173248.8
|
Ank1
|
ankyrin 1, erythroid |
| chr19_+_23881821 | 0.44 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr7_-_81216687 | 0.44 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr2_-_103858632 | 0.43 |
ENSMUST00000056170.4
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr16_-_13977084 | 0.43 |
ENSMUST00000090300.6
|
Marf1
|
meiosis regulator and mRNA stability 1 |
| chr3_+_84500854 | 0.43 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr13_-_74512111 | 0.42 |
ENSMUST00000022064.5
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr13_+_102830104 | 0.41 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr10_+_127256736 | 0.40 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr6_-_142647985 | 0.40 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_+_6047112 | 0.39 |
ENSMUST00000115786.8
|
Synj2
|
synaptojanin 2 |
| chr1_+_131671751 | 0.39 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr2_-_77000936 | 0.38 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr15_+_25940859 | 0.38 |
ENSMUST00000226750.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr11_+_46126274 | 0.37 |
ENSMUST00000060185.3
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr6_+_53264255 | 0.37 |
ENSMUST00000203528.3
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr2_+_118493713 | 0.36 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr2_-_151815307 | 0.36 |
ENSMUST00000109863.2
|
Fam110a
|
family with sequence similarity 110, member A |
| chr16_+_44913974 | 0.36 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr4_+_135975243 | 0.36 |
ENSMUST00000102533.11
ENSMUST00000143942.2 |
Tcea3
|
transcription elongation factor A (SII), 3 |
| chr1_+_42734051 | 0.35 |
ENSMUST00000239323.2
ENSMUST00000199521.5 ENSMUST00000176807.3 |
Pou3f3
Gm20646
|
POU domain, class 3, transcription factor 3 predicted gene 20646 |
| chr4_-_126150066 | 0.34 |
ENSMUST00000122129.8
ENSMUST00000061143.15 ENSMUST00000106132.3 |
Map7d1
|
MAP7 domain containing 1 |
| chr12_+_24701273 | 0.34 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr9_-_18675292 | 0.33 |
ENSMUST00000216754.3
|
Olfr24
|
olfactory receptor 24 |
| chr9_+_99511876 | 0.33 |
ENSMUST00000112886.9
|
Dzip1l
|
DAZ interacting protein 1-like |
| chr2_-_57014015 | 0.32 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr5_-_51711204 | 0.32 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr15_+_25940931 | 0.32 |
ENSMUST00000110438.3
|
Retreg1
|
reticulophagy regulator 1 |
| chr1_+_16758731 | 0.31 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr13_+_83672389 | 0.31 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_95603494 | 0.31 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr16_+_13176238 | 0.30 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr6_+_126992549 | 0.29 |
ENSMUST00000000187.7
|
Fgf6
|
fibroblast growth factor 6 |
| chr14_-_36641470 | 0.28 |
ENSMUST00000182042.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr10_+_127256993 | 0.28 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr1_+_132996237 | 0.28 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr2_+_102488985 | 0.27 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr9_-_70048766 | 0.27 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr19_-_40365318 | 0.26 |
ENSMUST00000239304.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_-_152642032 | 0.26 |
ENSMUST00000111859.8
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr7_+_142052569 | 0.25 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr16_-_36810810 | 0.25 |
ENSMUST00000075869.13
|
Fbxo40
|
F-box protein 40 |
| chr11_-_69260203 | 0.25 |
ENSMUST00000092971.13
ENSMUST00000108661.8 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr17_+_50816296 | 0.25 |
ENSMUST00000043938.8
|
Plcl2
|
phospholipase C-like 2 |
| chr6_+_134958681 | 0.25 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr1_-_135513443 | 0.24 |
ENSMUST00000067414.13
|
Nav1
|
neuron navigator 1 |
| chr5_+_64316771 | 0.24 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr11_+_73051228 | 0.24 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_+_11352862 | 0.23 |
ENSMUST00000188995.2
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr18_+_61096597 | 0.23 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr15_-_66432938 | 0.22 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr19_+_3758285 | 0.22 |
ENSMUST00000237320.2
ENSMUST00000039048.2 ENSMUST00000235295.2 ENSMUST00000237955.2 ENSMUST00000235837.2 |
1810055G02Rik
|
RIKEN cDNA 1810055G02 gene |
| chr4_+_11485945 | 0.22 |
ENSMUST00000055372.14
ENSMUST00000059914.13 |
Virma
|
vir like m6A methyltransferase associated |
| chr15_+_25940781 | 0.22 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr17_+_71326510 | 0.22 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr7_+_28510350 | 0.22 |
ENSMUST00000174477.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_-_113166153 | 0.21 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr18_-_47501897 | 0.21 |
ENSMUST00000019791.14
ENSMUST00000115449.9 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_-_186438177 | 0.21 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr7_-_115630282 | 0.20 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_156300325 | 0.19 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr6_+_121277186 | 0.19 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr16_-_90866032 | 0.19 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr11_+_67090878 | 0.18 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr16_+_43184191 | 0.18 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_-_30755007 | 0.18 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr11_-_69838971 | 0.17 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr2_-_92222979 | 0.17 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr4_+_133280680 | 0.16 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr10_+_22520910 | 0.16 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chrX_+_6489313 | 0.16 |
ENSMUST00000089520.3
|
Shroom4
|
shroom family member 4 |
| chr19_+_21249636 | 0.16 |
ENSMUST00000237651.2
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr18_+_69652751 | 0.16 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr6_-_116050081 | 0.15 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr13_-_103233323 | 0.15 |
ENSMUST00000166336.9
ENSMUST00000239261.2 ENSMUST00000194446.7 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_75351914 | 0.15 |
ENSMUST00000087122.12
|
Speg
|
SPEG complex locus |
| chr4_+_46039202 | 0.15 |
ENSMUST00000156200.8
|
Tmod1
|
tropomodulin 1 |
| chr5_+_122239007 | 0.15 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr7_-_101486983 | 0.15 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr12_-_109566764 | 0.15 |
ENSMUST00000149046.4
|
Rtl1
|
retrotransposon Gaglike 1 |
| chr14_-_66518399 | 0.14 |
ENSMUST00000111121.2
ENSMUST00000022622.14 ENSMUST00000089250.9 |
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta |
| chr10_+_88036947 | 0.13 |
ENSMUST00000020248.16
ENSMUST00000182183.8 ENSMUST00000171151.9 ENSMUST00000182619.2 |
Washc3
|
WASH complex subunit 3 |
| chr8_-_41469786 | 0.13 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chrX_-_58613428 | 0.12 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chrX_-_161747552 | 0.12 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr18_-_79152504 | 0.12 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr17_+_85335775 | 0.11 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2d_Mef2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.6 | 2.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.5 | 3.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.4 | 1.1 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.3 | 3.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 1.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 1.6 | GO:1904048 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.2 | 1.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 0.7 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.2 | 0.9 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 1.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 2.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 2.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.9 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 2.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.7 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.4 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.8 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.7 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 1.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.5 | GO:2000224 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.1 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.0 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.0 | 0.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 3.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 3.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.9 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 2.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 2.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.6 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.7 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 1.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 3.5 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0000802 | transverse filament(GO:0000802) nuclear meiotic cohesin complex(GO:0034991) |
| 0.4 | 1.1 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.2 | 1.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.2 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 1.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.7 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 2.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 5.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.1 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.6 | 1.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.3 | 1.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.6 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 1.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 3.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 4.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |