Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Meox2
Z-value: 0.60
Transcription factors associated with Meox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox2
|
ENSMUSG00000036144.7 | mesenchyme homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox2 | mm39_v1_chr12_+_37158532_37158545 | 0.69 | 2.0e-01 | Click! |
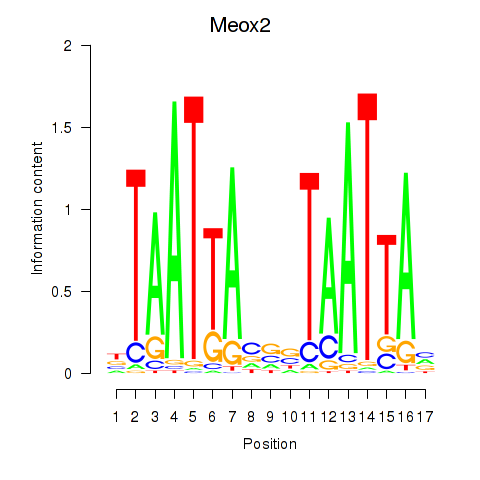
Activity profile of Meox2 motif
Sorted Z-values of Meox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23755551 | 0.68 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr7_-_3901119 | 0.64 |
ENSMUST00000070639.8
|
Gm14548
|
predicted gene 14548 |
| chr15_+_31224555 | 0.58 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr10_+_129072073 | 0.52 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chrM_+_9870 | 0.52 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr16_-_58620631 | 0.49 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr13_-_23755374 | 0.43 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr7_-_3828640 | 0.42 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr9_-_19799300 | 0.41 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr8_+_66838927 | 0.40 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chrM_-_14061 | 0.37 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrM_+_14138 | 0.37 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr16_+_65317389 | 0.36 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr11_+_73851643 | 0.31 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr7_+_100435548 | 0.31 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr5_+_109567554 | 0.31 |
ENSMUST00000232833.2
ENSMUST00000232722.2 ENSMUST00000233536.2 ENSMUST00000171841.3 ENSMUST00000233038.2 ENSMUST00000233724.2 |
Vmn2r17
|
vomeronasal 2, receptor 17 |
| chr9_+_20193647 | 0.31 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr2_+_88285211 | 0.30 |
ENSMUST00000219871.2
ENSMUST00000213115.3 |
Olfr1183
|
olfactory receptor 1183 |
| chr9_+_38164070 | 0.29 |
ENSMUST00000213129.2
|
Olfr143
|
olfactory receptor 143 |
| chr5_-_109372574 | 0.28 |
ENSMUST00000170341.3
|
Vmn2r14
|
vomeronasal 2, receptor 14 |
| chr11_-_71092282 | 0.28 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr5_-_108017019 | 0.27 |
ENSMUST00000128723.8
ENSMUST00000124034.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr6_+_56974228 | 0.27 |
ENSMUST00000228285.2
ENSMUST00000227847.2 ENSMUST00000226689.2 ENSMUST00000227131.2 ENSMUST00000227631.2 ENSMUST00000227188.2 |
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr16_-_55659194 | 0.27 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr13_-_67454476 | 0.27 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr16_+_57369595 | 0.27 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_-_10292412 | 0.25 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr9_+_38312994 | 0.24 |
ENSMUST00000214648.3
ENSMUST00000056364.3 |
Olfr147
|
olfactory receptor 147 |
| chr17_+_17924009 | 0.24 |
ENSMUST00000169805.3
|
Vmn2r90
|
vomeronasal 2, receptor 90 |
| chr2_-_87838612 | 0.23 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr2_-_26184450 | 0.23 |
ENSMUST00000217256.2
ENSMUST00000227200.2 |
Ccdc187
|
coiled-coil domain containing 187 |
| chrM_+_3906 | 0.23 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr9_-_39618413 | 0.22 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149 |
| chr2_-_111779785 | 0.22 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr13_-_30039613 | 0.22 |
ENSMUST00000091674.12
ENSMUST00000006353.14 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr18_+_60936910 | 0.21 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr2_-_86070633 | 0.21 |
ENSMUST00000215607.3
|
Olfr1048
|
olfactory receptor 1048 |
| chr2_+_120459593 | 0.19 |
ENSMUST00000180041.9
|
Stard9
|
START domain containing 9 |
| chr2_-_86926352 | 0.19 |
ENSMUST00000217066.3
ENSMUST00000214636.2 |
Olfr1109
|
olfactory receptor 1109 |
| chr2_+_87696836 | 0.19 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr9_+_39367997 | 0.19 |
ENSMUST00000214818.3
|
Olfr954
|
olfactory receptor 954 |
| chr2_-_45000389 | 0.18 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_37007795 | 0.18 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr2_-_45000250 | 0.18 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr16_-_59138611 | 0.18 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr5_+_107977965 | 0.17 |
ENSMUST00000072578.8
|
Ube2d2b
|
ubiquitin-conjugating enzyme E2D 2B |
| chr17_+_21446349 | 0.17 |
ENSMUST00000235895.2
|
Vmn1r234
|
vomeronasal 1 receptor 234 |
| chr19_-_12302465 | 0.17 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chr6_-_68609426 | 0.17 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr9_+_38053813 | 0.17 |
ENSMUST00000213458.2
|
Olfr890
|
olfactory receptor 890 |
| chrM_+_11735 | 0.16 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_-_52859227 | 0.14 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr2_+_32617671 | 0.14 |
ENSMUST00000113242.5
|
Sh2d3c
|
SH2 domain containing 3C |
| chr1_+_174000304 | 0.14 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr7_-_102507962 | 0.14 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr15_+_31225302 | 0.14 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr2_-_88157559 | 0.14 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr10_-_88518878 | 0.14 |
ENSMUST00000004473.15
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr8_+_3410842 | 0.13 |
ENSMUST00000238813.2
|
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr1_-_194859015 | 0.13 |
ENSMUST00000082321.9
ENSMUST00000195120.6 |
Cr2
|
complement receptor 2 |
| chr7_+_5221492 | 0.13 |
ENSMUST00000228062.2
ENSMUST00000227798.2 |
Vmn1r57
|
vomeronasal 1 receptor 57 |
| chr11_-_71092124 | 0.13 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr18_+_75133519 | 0.13 |
ENSMUST00000079716.6
|
Rpl17
|
ribosomal protein L17 |
| chr2_+_136733421 | 0.13 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr18_+_37580692 | 0.12 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr9_+_38202403 | 0.12 |
ENSMUST00000216276.2
ENSMUST00000215219.2 |
Olfr896-ps1
|
olfactory receptor 896, pseudogene 1 |
| chrM_+_10167 | 0.12 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_-_152687877 | 0.12 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr6_+_57043772 | 0.11 |
ENSMUST00000228714.2
ENSMUST00000227399.2 |
Vmn1r9
|
vomeronasal 1 receptor 9 |
| chr14_+_50360643 | 0.11 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr9_+_19247753 | 0.11 |
ENSMUST00000215572.3
ENSMUST00000213344.2 |
Olfr845
|
olfactory receptor 845 |
| chr7_-_34012956 | 0.11 |
ENSMUST00000108074.8
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr9_-_19289965 | 0.11 |
ENSMUST00000216839.3
|
Olfr847
|
olfactory receptor 847 |
| chr12_-_114451189 | 0.11 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr7_-_34012934 | 0.11 |
ENSMUST00000206399.2
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr4_+_134658209 | 0.11 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr17_-_15784582 | 0.10 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr9_+_20209828 | 0.10 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr16_+_42727926 | 0.10 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_81102740 | 0.10 |
ENSMUST00000046114.5
|
Mrpl54
|
mitochondrial ribosomal protein L54 |
| chr5_-_109340065 | 0.10 |
ENSMUST00000053253.10
|
Vmn2r13
|
vomeronasal 2, receptor 13 |
| chr7_+_86444235 | 0.10 |
ENSMUST00000233099.2
ENSMUST00000164996.2 |
Vmn2r77
|
vomeronasal 2, receptor 77 |
| chr2_-_26299112 | 0.10 |
ENSMUST00000114090.8
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chrX_+_41157242 | 0.10 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr5_+_109478028 | 0.10 |
ENSMUST00000165180.3
ENSMUST00000232912.2 |
Vmn2r16
|
vomeronasal 2, receptor 16 |
| chr11_-_69556888 | 0.09 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr1_+_130754413 | 0.09 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr14_+_73376192 | 0.09 |
ENSMUST00000171070.8
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr16_-_57113207 | 0.09 |
ENSMUST00000023434.15
ENSMUST00000120112.2 ENSMUST00000119407.8 |
Tmem30c
|
transmembrane protein 30C |
| chr7_-_127897253 | 0.09 |
ENSMUST00000033044.16
|
Rusf1
|
RUS family member 1 |
| chr5_+_146885450 | 0.09 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr10_-_116808514 | 0.09 |
ENSMUST00000092165.5
|
Gm10271
|
predicted gene 10271 |
| chr11_+_74077075 | 0.09 |
ENSMUST00000206114.3
|
Olfr403
|
olfactory receptor 403 |
| chr17_+_20950105 | 0.08 |
ENSMUST00000233597.2
ENSMUST00000232943.2 ENSMUST00000179914.3 |
Vmn1r227
|
vomeronasal 1 receptor 227 |
| chr2_-_165230165 | 0.08 |
ENSMUST00000103084.4
|
Zfp334
|
zinc finger protein 334 |
| chr2_+_89821565 | 0.08 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr17_-_6129968 | 0.08 |
ENSMUST00000024570.6
ENSMUST00000097432.10 |
Serac1
|
serine active site containing 1 |
| chr19_-_13828056 | 0.08 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr12_-_115825934 | 0.08 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr8_+_57004125 | 0.07 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr16_-_56891711 | 0.07 |
ENSMUST00000067173.8
ENSMUST00000227043.2 |
Tmem45a2
|
transmembrane protein 45A2 |
| chrX_+_164953444 | 0.07 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr5_-_109445510 | 0.07 |
ENSMUST00000167133.3
ENSMUST00000233141.2 ENSMUST00000233813.2 ENSMUST00000233282.2 |
Vmn2r15
|
vomeronasal 2, receptor 15 |
| chr1_-_155910546 | 0.07 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr1_+_180878797 | 0.07 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr9_+_72345267 | 0.07 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr16_+_4457805 | 0.07 |
ENSMUST00000038770.4
|
Vasn
|
vasorin |
| chr4_+_66745803 | 0.07 |
ENSMUST00000048096.12
ENSMUST00000107365.3 |
Tlr4
|
toll-like receptor 4 |
| chr19_-_8796641 | 0.07 |
ENSMUST00000088092.6
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chrX_-_125723491 | 0.06 |
ENSMUST00000081074.5
|
4932411N23Rik
|
RIKEN cDNA 4932411N23 gene |
| chr6_+_68247469 | 0.06 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr18_-_9282754 | 0.06 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr12_-_114878652 | 0.06 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr7_-_5808444 | 0.06 |
ENSMUST00000075085.7
|
Vmn1r63
|
vomeronasal 1 receptor 63 |
| chr9_-_105973975 | 0.06 |
ENSMUST00000121963.3
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr9_+_38288382 | 0.05 |
ENSMUST00000214865.2
|
Olfr251
|
olfactory receptor 251 |
| chr2_-_89671899 | 0.05 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr6_-_69877961 | 0.05 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr8_-_37200051 | 0.05 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr14_+_51599213 | 0.05 |
ENSMUST00000162998.2
|
Gm7247
|
predicted gene 7247 |
| chr19_-_13827773 | 0.05 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr2_-_26299173 | 0.05 |
ENSMUST00000145701.8
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr1_-_155910567 | 0.04 |
ENSMUST00000141878.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr6_-_41012435 | 0.04 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr7_+_126376099 | 0.04 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr9_+_64142483 | 0.04 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr10_-_119948890 | 0.04 |
ENSMUST00000020449.12
|
Helb
|
helicase (DNA) B |
| chr4_-_24800890 | 0.04 |
ENSMUST00000108214.9
|
Klhl32
|
kelch-like 32 |
| chr17_+_34457868 | 0.03 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr6_-_30693675 | 0.03 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr5_-_147831610 | 0.03 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr12_-_115798038 | 0.03 |
ENSMUST00000103544.3
|
Ighv1-75
|
immunoglobulin heavy variable 1-75 |
| chr16_-_58583539 | 0.03 |
ENSMUST00000214139.2
|
Olfr172
|
olfactory receptor 172 |
| chr9_-_20247390 | 0.03 |
ENSMUST00000212943.4
ENSMUST00000086473.4 |
Olfr18
|
olfactory receptor 18 |
| chr19_+_13037974 | 0.03 |
ENSMUST00000213806.2
ENSMUST00000214695.2 ENSMUST00000217568.2 |
Olfr1454
|
olfactory receptor 1454 |
| chr1_-_92446383 | 0.03 |
ENSMUST00000062353.12
|
Olfr1414
|
olfactory receptor 1414 |
| chr11_+_46701619 | 0.03 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr7_+_6733684 | 0.03 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr1_-_162726234 | 0.03 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr16_+_14724485 | 0.03 |
ENSMUST00000090277.3
|
Efcab1
|
EF-hand calcium binding domain 1 |
| chr13_+_46822992 | 0.03 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr12_+_76464301 | 0.03 |
ENSMUST00000063977.9
|
Ppp1r36
|
protein phosphatase 1, regulatory subunit 36 |
| chr3_-_59009231 | 0.03 |
ENSMUST00000085040.5
|
Gpr171
|
G protein-coupled receptor 171 |
| chr5_-_125371162 | 0.03 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr7_+_102220649 | 0.03 |
ENSMUST00000216524.2
|
Olfr550
|
olfactory receptor 550 |
| chr4_+_63133639 | 0.03 |
ENSMUST00000036300.13
|
Col27a1
|
collagen, type XXVII, alpha 1 |
| chr14_+_57762191 | 0.03 |
ENSMUST00000089494.6
|
Il17d
|
interleukin 17D |
| chr6_+_34575435 | 0.03 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr14_-_61495934 | 0.03 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr19_-_13075176 | 0.03 |
ENSMUST00000208913.2
ENSMUST00000215229.2 |
Olfr1457
|
olfactory receptor 1457 |
| chr10_-_81335966 | 0.03 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr11_+_119493806 | 0.02 |
ENSMUST00000026671.13
|
Rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr8_+_114369838 | 0.02 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr10_+_129306867 | 0.02 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr12_+_80691275 | 0.02 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr2_+_85677374 | 0.02 |
ENSMUST00000215347.3
|
Olfr1020
|
olfactory receptor 1020 |
| chr3_+_108093645 | 0.02 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr4_+_110254858 | 0.02 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr10_-_80850712 | 0.02 |
ENSMUST00000126317.2
ENSMUST00000092285.10 ENSMUST00000117805.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_-_86257093 | 0.02 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr13_+_65660492 | 0.02 |
ENSMUST00000081471.3
|
Gm10139
|
predicted gene 10139 |
| chr12_-_81531847 | 0.02 |
ENSMUST00000166723.8
ENSMUST00000110340.9 ENSMUST00000168463.8 ENSMUST00000169124.2 ENSMUST00000002757.11 |
Cox16
|
cytochrome c oxidase assembly protein 16 |
| chr2_+_128705829 | 0.02 |
ENSMUST00000028864.3
|
Fbln7
|
fibulin 7 |
| chr1_-_173301492 | 0.02 |
ENSMUST00000169797.2
|
Ifi206
|
interferon activated gene 206 |
| chr17_-_66901568 | 0.02 |
ENSMUST00000024914.4
|
Themis3
|
thymocyte selection associated family member 3 |
| chr6_-_116561156 | 0.02 |
ENSMUST00000061723.6
|
Olfr215
|
olfactory receptor 215 |
| chr19_-_8796677 | 0.02 |
ENSMUST00000096751.11
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr12_-_114621406 | 0.02 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr12_-_114815280 | 0.01 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr1_+_178233640 | 0.01 |
ENSMUST00000027775.9
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr1_+_82978269 | 0.01 |
ENSMUST00000223439.2
|
Gm47959
|
predicted gene, 47959 |
| chr1_+_24216691 | 0.01 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr4_-_60455331 | 0.01 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr10_+_40225272 | 0.01 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr19_-_8796288 | 0.01 |
ENSMUST00000153281.2
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr6_+_116466052 | 0.01 |
ENSMUST00000203700.3
|
Olfr211
|
olfactory receptor 211 |
| chr7_-_66915756 | 0.01 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr2_+_85876205 | 0.01 |
ENSMUST00000213496.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr14_+_74878280 | 0.01 |
ENSMUST00000036653.5
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr1_-_9818601 | 0.01 |
ENSMUST00000057438.7
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr17_-_50600620 | 0.01 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr14_+_41765952 | 0.01 |
ENSMUST00000168972.2
ENSMUST00000064162.11 |
1700024B05Rik
|
RIKEN cDNA 1700024B05 gene |
| chr13_-_66375387 | 0.01 |
ENSMUST00000167981.3
|
Gm10772
|
predicted gene 10772 |
| chr5_-_109154393 | 0.01 |
ENSMUST00000079163.5
ENSMUST00000233053.2 |
Vmn2r10
|
vomeronasal 2, receptor 10 |
| chr7_-_108449643 | 0.01 |
ENSMUST00000216092.2
ENSMUST00000217279.2 |
Olfr516
|
olfactory receptor 516 |
| chr13_+_83652280 | 0.01 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr17_+_6130061 | 0.01 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr7_+_6733561 | 0.01 |
ENSMUST00000200535.6
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr6_-_69477770 | 0.01 |
ENSMUST00000197448.2
|
Igkv4-58
|
immunoglobulin kappa variable 4-58 |
| chr7_-_6733411 | 0.01 |
ENSMUST00000239104.2
ENSMUST00000051209.11 |
Peg3
|
paternally expressed 3 |
| chr5_+_9316097 | 0.01 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr3_+_79252668 | 0.01 |
ENSMUST00000164216.6
ENSMUST00000199658.2 |
Gm17359
|
predicted gene, 17359 |
| chr5_+_73164226 | 0.01 |
ENSMUST00000031127.11
ENSMUST00000201304.2 |
Slc10a4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
| chr10_-_51507527 | 0.01 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr13_+_59860096 | 0.01 |
ENSMUST00000165133.3
|
Spata31d1b
|
spermatogenesis associated 31 subfamily D, member 1B |
| chr14_-_20546848 | 0.01 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr7_-_32755975 | 0.01 |
ENSMUST00000182640.8
ENSMUST00000182673.2 |
Scgb2b17
|
secretoglobin, family 2B, member 17 |
| chr9_-_106448182 | 0.01 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr16_-_19079594 | 0.01 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr2_+_61634797 | 0.01 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.3 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.3 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.2 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 1.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |