Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
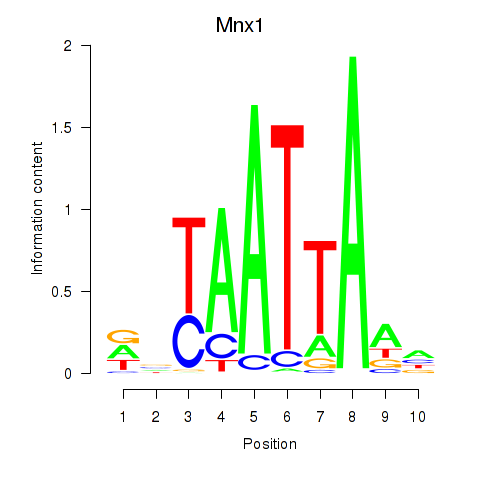
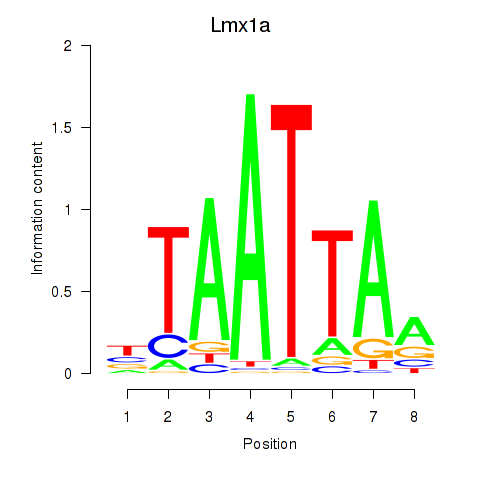
Results for Mnx1_Lhx6_Lmx1a
Z-value: 0.69
Transcription factors associated with Mnx1_Lhx6_Lmx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnx1
|
ENSMUSG00000001566.10 | motor neuron and pancreas homeobox 1 |
|
Lhx6
|
ENSMUSG00000026890.20 | LIM homeobox protein 6 |
|
Lmx1a
|
ENSMUSG00000026686.15 | LIM homeobox transcription factor 1 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnx1 | mm39_v1_chr5_-_29683468_29683595 | -0.86 | 6.2e-02 | Click! |
| Lhx6 | mm39_v1_chr2_-_35994072_35994085 | -0.80 | 1.0e-01 | Click! |
| Lmx1a | mm39_v1_chr1_+_167516771_167516806 | -0.80 | 1.1e-01 | Click! |
Activity profile of Mnx1_Lhx6_Lmx1a motif
Sorted Z-values of Mnx1_Lhx6_Lmx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_5815863 | 0.71 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr4_-_154721288 | 0.71 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chrM_+_2743 | 0.67 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr14_+_26722319 | 0.50 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr3_+_54268523 | 0.50 |
ENSMUST00000117373.8
ENSMUST00000107985.10 ENSMUST00000073012.13 ENSMUST00000081564.13 |
Postn
|
periostin, osteoblast specific factor |
| chrM_+_9870 | 0.49 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr15_-_8740218 | 0.45 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_129449739 | 0.39 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chrM_+_10167 | 0.38 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrM_+_9459 | 0.38 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr8_-_85389470 | 0.35 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr15_+_25774070 | 0.35 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr9_+_32027335 | 0.31 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_-_23821523 | 0.31 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr6_+_138117519 | 0.30 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr3_+_159545309 | 0.30 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chrM_+_7006 | 0.29 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_-_88317601 | 0.27 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr15_+_31224555 | 0.23 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr6_+_123239076 | 0.22 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr6_+_138117295 | 0.22 |
ENSMUST00000008684.11
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr7_-_108774367 | 0.21 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr11_-_3881995 | 0.19 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr18_+_84869456 | 0.18 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr5_-_65855511 | 0.18 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr11_-_3881960 | 0.18 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr10_+_128173603 | 0.17 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr19_-_24178000 | 0.17 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr11_-_3881789 | 0.17 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr12_-_25147139 | 0.17 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr13_-_43634695 | 0.17 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr7_-_45480200 | 0.17 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr15_+_65682066 | 0.17 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chrX_-_133012600 | 0.15 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr3_-_130524024 | 0.15 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr18_+_23885390 | 0.15 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_79650496 | 0.15 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr17_+_7437500 | 0.14 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr5_+_115373895 | 0.14 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr5_-_77262968 | 0.14 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr2_-_174188505 | 0.13 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr2_+_132689640 | 0.13 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr19_+_24853039 | 0.13 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr14_+_56003406 | 0.12 |
ENSMUST00000057569.4
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr14_-_64654397 | 0.12 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr10_-_129107354 | 0.12 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr3_+_106020545 | 0.12 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr18_-_43610829 | 0.12 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr15_+_31225302 | 0.11 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chrM_+_7779 | 0.11 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr11_-_87249837 | 0.11 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr16_-_92196954 | 0.11 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr7_-_44753168 | 0.11 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr15_+_31224616 | 0.11 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr3_-_50398027 | 0.11 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr18_+_23548534 | 0.11 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr9_-_63929308 | 0.11 |
ENSMUST00000179458.2
ENSMUST00000041029.6 |
Smad6
|
SMAD family member 6 |
| chr14_+_32507920 | 0.11 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr7_-_99512558 | 0.11 |
ENSMUST00000207137.2
ENSMUST00000207063.2 ENSMUST00000207580.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr19_-_15902292 | 0.11 |
ENSMUST00000025542.10
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr9_+_113641615 | 0.10 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chrM_+_11735 | 0.10 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr8_-_85573489 | 0.10 |
ENSMUST00000003912.7
|
Calr
|
calreticulin |
| chr2_+_36120438 | 0.10 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_-_138169253 | 0.10 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr6_+_83142902 | 0.10 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr8_-_32440071 | 0.10 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chr7_-_24423715 | 0.10 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr11_+_23256883 | 0.10 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr3_+_144283355 | 0.10 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F |
| chr19_-_15901919 | 0.10 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr14_+_80237691 | 0.10 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr12_+_70499869 | 0.10 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr18_+_23548455 | 0.09 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr15_-_11038077 | 0.09 |
ENSMUST00000058007.7
|
Rxfp3
|
relaxin family peptide receptor 3 |
| chr14_-_64654592 | 0.09 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr11_-_99265721 | 0.09 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr4_+_102848981 | 0.09 |
ENSMUST00000140654.9
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr9_+_65797519 | 0.09 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr5_-_137530214 | 0.09 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr10_-_35587888 | 0.08 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr3_-_106697459 | 0.08 |
ENSMUST00000038845.10
|
Cd53
|
CD53 antigen |
| chr13_-_103042554 | 0.08 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_20742115 | 0.08 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr11_-_120440519 | 0.08 |
ENSMUST00000034913.5
|
Mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr3_+_40755211 | 0.08 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr14_+_55797468 | 0.08 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_-_79658608 | 0.08 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr1_-_69726384 | 0.08 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr2_+_83554741 | 0.07 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr3_-_49711765 | 0.07 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr1_+_179788037 | 0.07 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr4_-_14621805 | 0.07 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr8_-_62576140 | 0.07 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr10_-_53252210 | 0.07 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr2_-_154916367 | 0.07 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr9_+_62765362 | 0.07 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr4_+_108576846 | 0.07 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr4_+_148985584 | 0.07 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chr8_-_62355690 | 0.07 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr10_-_20424069 | 0.07 |
ENSMUST00000169404.8
|
Pde7b
|
phosphodiesterase 7B |
| chr7_-_84339045 | 0.07 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr3_-_49711706 | 0.07 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr14_+_55797443 | 0.07 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr10_+_23727325 | 0.07 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr3_+_55689921 | 0.07 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr9_+_108216466 | 0.07 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr14_+_79753055 | 0.07 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr8_-_79539838 | 0.06 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_-_137529465 | 0.06 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr15_+_21111428 | 0.06 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr3_+_68479578 | 0.06 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_-_72323464 | 0.06 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr3_-_130523954 | 0.06 |
ENSMUST00000196202.5
ENSMUST00000133802.6 ENSMUST00000062601.14 ENSMUST00000200517.2 |
Rpl34
|
ribosomal protein L34 |
| chr7_+_25380195 | 0.06 |
ENSMUST00000205658.2
|
B9d2
|
B9 protein domain 2 |
| chr10_+_87926932 | 0.06 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_-_27365633 | 0.06 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr5_+_43390513 | 0.06 |
ENSMUST00000166713.9
ENSMUST00000169035.8 ENSMUST00000114065.9 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr3_+_67799510 | 0.06 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr3_-_86455575 | 0.06 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr18_+_24737009 | 0.06 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr4_-_35845204 | 0.06 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_+_88234454 | 0.06 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr9_-_71070506 | 0.06 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr16_-_89615225 | 0.06 |
ENSMUST00000164263.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr2_+_89642395 | 0.06 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr6_+_71886030 | 0.06 |
ENSMUST00000055296.11
|
Polr1a
|
polymerase (RNA) I polypeptide A |
| chr10_+_79746690 | 0.06 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr11_+_60428788 | 0.05 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chrX_-_142716085 | 0.05 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr10_-_107321938 | 0.05 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr10_+_85222677 | 0.05 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr10_-_53952686 | 0.05 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr1_+_171668173 | 0.05 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr2_-_5838489 | 0.05 |
ENSMUST00000128467.4
|
Cdc123
|
cell division cycle 123 |
| chr5_+_35156389 | 0.05 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_+_126550009 | 0.05 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr17_+_38104420 | 0.05 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr19_-_11582207 | 0.05 |
ENSMUST00000025582.11
|
Ms4a6d
|
membrane-spanning 4-domains, subfamily A, member 6D |
| chr12_-_40087393 | 0.05 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr7_+_25380263 | 0.05 |
ENSMUST00000205325.2
ENSMUST00000206913.2 |
B9d2
|
B9 protein domain 2 |
| chr3_+_32490300 | 0.05 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr1_-_182929025 | 0.05 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr9_+_89081407 | 0.05 |
ENSMUST00000138109.2
|
Gm29094
|
predicted gene 29094 |
| chr12_+_103524690 | 0.05 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr18_-_57108405 | 0.05 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr9_+_118307412 | 0.05 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr2_-_86109346 | 0.05 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr2_+_87686206 | 0.05 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr16_-_74208180 | 0.05 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr3_-_89257266 | 0.05 |
ENSMUST00000107446.8
ENSMUST00000074582.7 ENSMUST00000107448.9 ENSMUST00000029676.12 |
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin) |
| chr7_-_37718916 | 0.05 |
ENSMUST00000085513.6
ENSMUST00000206327.2 |
Uri1
|
URI1, prefoldin-like chaperone |
| chr8_+_107757847 | 0.05 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr16_+_19578981 | 0.05 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr2_-_18053595 | 0.05 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr2_-_5680801 | 0.04 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr9_+_43222104 | 0.04 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr18_+_9707595 | 0.04 |
ENSMUST00000234965.2
|
Colec12
|
collectin sub-family member 12 |
| chr5_+_20112500 | 0.04 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_143864993 | 0.04 |
ENSMUST00000172367.3
|
Gm42421
|
predicted gene, 42421 |
| chr5_+_138185747 | 0.04 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr1_-_171854818 | 0.04 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr18_+_31742565 | 0.04 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr17_+_38104635 | 0.04 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr5_+_38233901 | 0.04 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr19_-_41921676 | 0.04 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr7_-_10488291 | 0.04 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr7_-_101486983 | 0.04 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr2_-_168609110 | 0.04 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr2_+_85612365 | 0.04 |
ENSMUST00000215945.2
|
Olfr1015
|
olfactory receptor 1015 |
| chr19_-_50667079 | 0.04 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr11_-_6217718 | 0.04 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
| chr18_+_23886765 | 0.04 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr8_-_32408864 | 0.04 |
ENSMUST00000073884.7
ENSMUST00000238812.2 |
Nrg1
|
neuregulin 1 |
| chr10_-_6930376 | 0.04 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_-_22289994 | 0.04 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr2_-_87504008 | 0.04 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr8_-_68270870 | 0.04 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_93373364 | 0.04 |
ENSMUST00000190321.7
ENSMUST00000042498.14 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr11_-_99494134 | 0.04 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr5_-_86437119 | 0.04 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr13_-_103042294 | 0.04 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_87435091 | 0.04 |
ENSMUST00000021424.5
|
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr11_+_23256909 | 0.04 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr5_+_9150691 | 0.04 |
ENSMUST00000115365.3
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr13_+_24986003 | 0.04 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr6_-_50631418 | 0.04 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr19_+_58500411 | 0.04 |
ENSMUST00000235305.2
ENSMUST00000069419.8 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr1_-_72323407 | 0.04 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr7_+_107411470 | 0.03 |
ENSMUST00000208563.3
ENSMUST00000214253.2 |
Olfr467
|
olfactory receptor 467 |
| chr15_-_79658584 | 0.03 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr17_-_71160477 | 0.03 |
ENSMUST00000118283.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_-_107228382 | 0.03 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr19_-_55229668 | 0.03 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr4_-_14621497 | 0.03 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_+_35156454 | 0.03 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr11_-_59466995 | 0.03 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr18_-_24736521 | 0.03 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr10_-_20424101 | 0.03 |
ENSMUST00000164195.2
|
Pde7b
|
phosphodiesterase 7B |
| chr19_+_41921903 | 0.03 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnx1_Lhx6_Lmx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.5 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.1 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.8 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:1902867 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.0 | 1.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.0 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.0 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.0 | 0.0 | GO:1904753 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 1.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |