Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
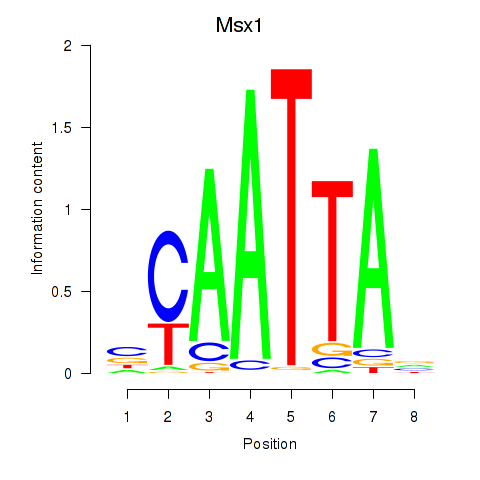
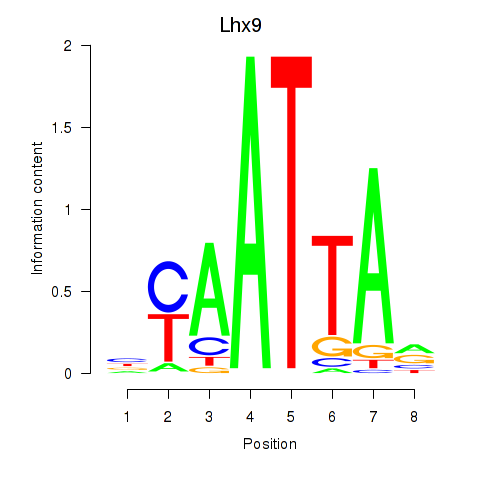
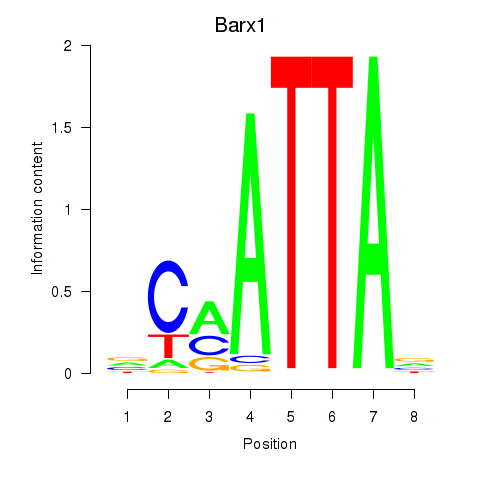
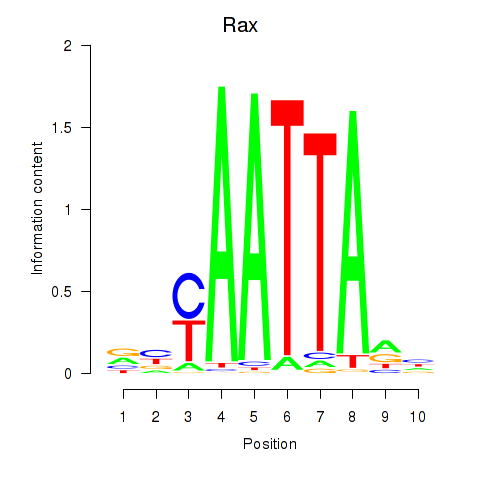
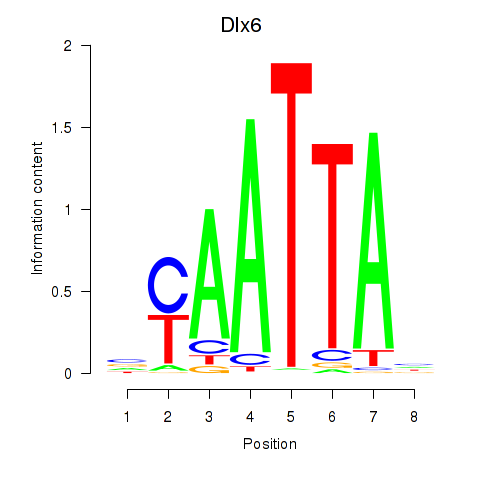
Results for Msx1_Lhx9_Barx1_Rax_Dlx6
Z-value: 0.27
Transcription factors associated with Msx1_Lhx9_Barx1_Rax_Dlx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx1
|
ENSMUSG00000048450.11 | msh homeobox 1 |
|
Lhx9
|
ENSMUSG00000019230.15 | LIM homeobox protein 9 |
|
Barx1
|
ENSMUSG00000021381.5 | BarH-like homeobox 1 |
|
Rax
|
ENSMUSG00000024518.5 | retina and anterior neural fold homeobox |
|
Dlx6
|
ENSMUSG00000029754.14 | distal-less homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx6 | mm39_v1_chr6_+_6863269_6863334 | -0.98 | 2.3e-03 | Click! |
| Lhx9 | mm39_v1_chr1_-_138776315_138776341 | -0.97 | 5.1e-03 | Click! |
| Rax | mm39_v1_chr18_-_66072119_66072160 | -0.90 | 3.7e-02 | Click! |
| Msx1 | mm39_v1_chr5_-_37981919_37981933 | -0.76 | 1.4e-01 | Click! |
| Barx1 | mm39_v1_chr13_+_48816466_48816521 | 0.30 | 6.2e-01 | Click! |
Activity profile of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
Sorted Z-values of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_99099084 | 0.34 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr3_+_159545309 | 0.21 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr19_-_24178000 | 0.21 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr5_-_77262968 | 0.20 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr14_+_26722319 | 0.18 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_+_115373895 | 0.15 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr18_-_43610829 | 0.15 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_-_45480200 | 0.14 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr14_+_79753055 | 0.14 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr10_+_127257077 | 0.12 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr9_+_118307412 | 0.12 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr6_-_129449739 | 0.10 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr15_+_65682066 | 0.09 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr5_+_35156389 | 0.09 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr3_+_68479578 | 0.09 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr15_+_21111428 | 0.09 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr18_-_66155651 | 0.09 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr5_-_23821523 | 0.08 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr15_+_25774070 | 0.08 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr5_-_137530214 | 0.08 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_+_60428788 | 0.08 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr19_+_26727111 | 0.07 |
ENSMUST00000175842.4
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrM_+_7006 | 0.07 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr11_-_87249837 | 0.07 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr2_+_132689640 | 0.07 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr9_+_32027335 | 0.07 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_-_4909515 | 0.06 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr12_-_25147139 | 0.06 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr8_+_107757847 | 0.06 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr13_-_43634695 | 0.06 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chrM_+_8603 | 0.06 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_+_23256883 | 0.06 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chrX_+_158086253 | 0.06 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr7_+_126550009 | 0.06 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr14_-_86986541 | 0.06 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr5_-_137529465 | 0.05 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr12_+_70499869 | 0.05 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr17_+_46471950 | 0.05 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr5_-_65855511 | 0.05 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr8_-_85389470 | 0.05 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr18_+_24737009 | 0.05 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr11_+_23256909 | 0.05 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr3_+_151143524 | 0.05 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr6_+_29859372 | 0.05 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr5_+_35156454 | 0.05 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_-_138185438 | 0.05 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr2_+_170353338 | 0.05 |
ENSMUST00000136839.2
ENSMUST00000109148.8 ENSMUST00000170167.8 |
Pfdn4
|
prefoldin 4 |
| chr10_-_107321938 | 0.05 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr9_-_103099262 | 0.04 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr16_-_95387444 | 0.04 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor |
| chr18_+_84869456 | 0.04 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr11_-_99265721 | 0.04 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr8_-_85573489 | 0.04 |
ENSMUST00000003912.7
|
Calr
|
calreticulin |
| chr17_+_45817750 | 0.04 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr18_+_23548455 | 0.04 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr3_+_96537484 | 0.04 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr10_+_82695013 | 0.04 |
ENSMUST00000020484.9
ENSMUST00000219962.3 |
Txnrd1
|
thioredoxin reductase 1 |
| chr11_-_107228382 | 0.04 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr15_-_8740218 | 0.04 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_+_102175985 | 0.04 |
ENSMUST00000156326.2
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_-_52230062 | 0.04 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr16_-_92196954 | 0.04 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr9_-_71070506 | 0.04 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr2_+_20742115 | 0.04 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr13_-_103042554 | 0.04 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_-_49711765 | 0.04 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr1_-_171854818 | 0.03 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr7_+_101545547 | 0.03 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr12_+_55286111 | 0.03 |
ENSMUST00000164243.2
|
Srp54c
|
signal recognition particle 54C |
| chr3_-_49711706 | 0.03 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr10_+_127919142 | 0.03 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr2_-_5838489 | 0.03 |
ENSMUST00000128467.4
|
Cdc123
|
cell division cycle 123 |
| chr7_-_44635740 | 0.03 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr17_+_27775613 | 0.03 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr1_-_4430481 | 0.03 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_+_71219561 | 0.03 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr7_+_101546059 | 0.03 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr18_+_23548534 | 0.03 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr1_+_11063678 | 0.03 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr12_+_38830283 | 0.03 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chrX_+_41241049 | 0.03 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr18_-_57108405 | 0.03 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr16_+_35803794 | 0.03 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr17_+_38104420 | 0.03 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr2_-_69542805 | 0.03 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr18_-_38102799 | 0.03 |
ENSMUST00000166148.8
ENSMUST00000163131.8 ENSMUST00000043437.14 |
Fchsd1
|
FCH and double SH3 domains 1 |
| chr17_+_27775637 | 0.03 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr5_-_123126550 | 0.03 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_-_97944239 | 0.03 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr6_-_52203146 | 0.03 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr14_-_52341472 | 0.03 |
ENSMUST00000111610.12
ENSMUST00000164655.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr12_-_91815855 | 0.03 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr4_-_14621805 | 0.03 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_+_89642395 | 0.03 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr5_+_138185747 | 0.03 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr5_-_138185686 | 0.02 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr3_-_88317601 | 0.02 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr18_+_56565188 | 0.02 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr9_-_40257586 | 0.02 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr12_+_72488625 | 0.02 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr17_+_38104635 | 0.02 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr11_-_99494134 | 0.02 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr17_+_35150229 | 0.02 |
ENSMUST00000007253.6
|
Neu1
|
neuraminidase 1 |
| chr10_+_18345706 | 0.02 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr3_+_96537235 | 0.02 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr11_-_99228756 | 0.02 |
ENSMUST00000100482.3
|
Krt26
|
keratin 26 |
| chr6_-_99243455 | 0.02 |
ENSMUST00000113326.9
|
Foxp1
|
forkhead box P1 |
| chr18_-_81029986 | 0.02 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr2_-_77533596 | 0.02 |
ENSMUST00000171063.8
|
Zfp385b
|
zinc finger protein 385B |
| chr19_+_45433899 | 0.02 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr12_+_38830812 | 0.02 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr5_-_86437119 | 0.02 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr12_-_21467437 | 0.02 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr9_-_96513529 | 0.02 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr2_+_85612365 | 0.02 |
ENSMUST00000215945.2
|
Olfr1015
|
olfactory receptor 1015 |
| chr5_-_123127346 | 0.02 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr9_-_79884920 | 0.02 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr2_-_168608949 | 0.02 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr9_-_79885063 | 0.02 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr10_-_8632519 | 0.02 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr14_-_36820304 | 0.02 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr2_+_69050315 | 0.02 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr2_+_127750978 | 0.02 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr15_-_34356567 | 0.02 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr17_+_34258411 | 0.02 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr7_+_126549859 | 0.02 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr19_-_55229668 | 0.02 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr6_+_15196950 | 0.02 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr2_+_27055245 | 0.02 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr18_+_37898633 | 0.02 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr4_-_14621497 | 0.02 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_+_30463175 | 0.02 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr4_+_150938376 | 0.02 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr15_+_28203872 | 0.02 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr3_+_67799510 | 0.02 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr14_-_54651442 | 0.01 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr10_-_129107354 | 0.01 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr7_-_115797722 | 0.01 |
ENSMUST00000181981.8
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr3_+_151143557 | 0.01 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr8_+_10299288 | 0.01 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr7_-_44635813 | 0.01 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr16_+_35861554 | 0.01 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr10_+_82695055 | 0.01 |
ENSMUST00000218694.2
|
Txnrd1
|
thioredoxin reductase 1 |
| chr7_+_107411470 | 0.01 |
ENSMUST00000208563.3
ENSMUST00000214253.2 |
Olfr467
|
olfactory receptor 467 |
| chr13_-_103042294 | 0.01 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_165762469 | 0.01 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr11_-_100653754 | 0.01 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr14_-_52341426 | 0.01 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr2_-_87504008 | 0.01 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr9_+_43222104 | 0.01 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr14_-_110992533 | 0.01 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr19_+_26728339 | 0.01 |
ENSMUST00000175953.3
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_108000425 | 0.01 |
ENSMUST00000151326.8
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr11_-_20282684 | 0.01 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr10_+_73782857 | 0.01 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr10_+_26648473 | 0.01 |
ENSMUST00000039557.9
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr5_-_123127148 | 0.01 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_+_98689479 | 0.01 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chr2_-_168609110 | 0.01 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr6_-_50631418 | 0.01 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr11_-_77079794 | 0.01 |
ENSMUST00000108400.8
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr11_-_115167775 | 0.01 |
ENSMUST00000021078.3
|
Fdxr
|
ferredoxin reductase |
| chr8_+_120955195 | 0.01 |
ENSMUST00000180448.3
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr17_-_52139693 | 0.01 |
ENSMUST00000144331.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr15_-_101801351 | 0.01 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr2_-_27365633 | 0.01 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr1_-_173018204 | 0.01 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr19_-_46033353 | 0.01 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr1_-_79417732 | 0.01 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr6_+_29859685 | 0.01 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr2_-_86109346 | 0.01 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr3_+_66127330 | 0.01 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr11_+_99748741 | 0.01 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr6_+_29859660 | 0.01 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chrX_-_94701983 | 0.01 |
ENSMUST00000119640.8
ENSMUST00000120620.8 ENSMUST00000044382.7 |
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr14_-_118289557 | 0.01 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr2_+_73102269 | 0.01 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr4_+_19280850 | 0.01 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr14_+_32043944 | 0.01 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr6_+_63232955 | 0.01 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr4_-_35845204 | 0.01 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chrX_+_55500170 | 0.01 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr5_+_14075281 | 0.01 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr2_+_85648823 | 0.01 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr16_-_63684477 | 0.01 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr8_+_22996233 | 0.01 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr4_+_21848039 | 0.01 |
ENSMUST00000098238.9
ENSMUST00000108229.2 |
Pnisr
|
PNN interacting serine/arginine-rich |
| chr3_-_141687987 | 0.01 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr10_+_73657753 | 0.01 |
ENSMUST00000134009.9
ENSMUST00000177420.7 ENSMUST00000125006.9 |
Pcdh15
|
protocadherin 15 |
| chr8_-_62576140 | 0.01 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr6_-_136148820 | 0.01 |
ENSMUST00000188999.3
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr10_-_80257681 | 0.01 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr3_+_55689921 | 0.01 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr12_-_104439589 | 0.01 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr17_-_31417834 | 0.01 |
ENSMUST00000236793.2
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr7_-_115459082 | 0.01 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr18_-_81029751 | 0.01 |
ENSMUST00000238808.2
|
Sall3
|
spalt like transcription factor 3 |
| chr11_+_102495189 | 0.01 |
ENSMUST00000057893.7
|
Fzd2
|
frizzled class receptor 2 |
| chr6_+_28480336 | 0.01 |
ENSMUST00000001460.14
ENSMUST00000167201.2 |
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr12_+_38831093 | 0.01 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx1_Lhx9_Barx1_Rax_Dlx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |