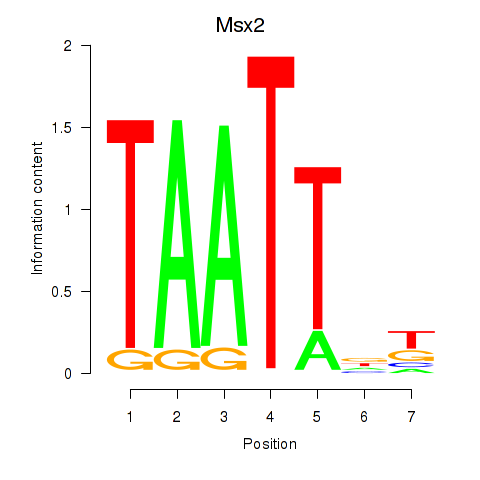
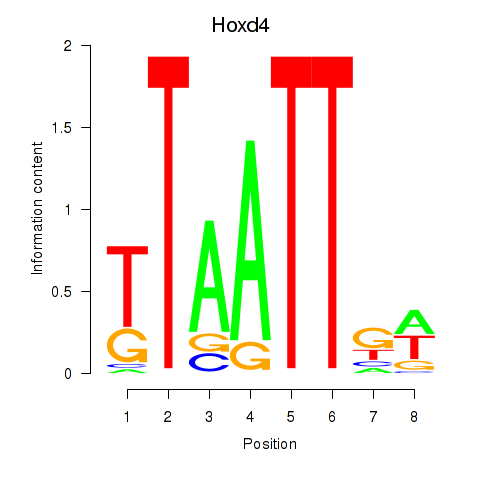
|
chr13_+_23728222
Show fit
|
1.15 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7
|
|
chr5_-_62923463
Show fit
|
0.99 |
ENSMUST00000076623.8
ENSMUST00000159470.3
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr2_-_104324035
Show fit
|
0.77 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3
|
|
chr5_+_107655487
Show fit
|
0.67 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669
|
|
chr7_+_140181182
Show fit
|
0.60 |
ENSMUST00000214180.2
ENSMUST00000211771.2
|
Olfr46
|
olfactory receptor 46
|
|
chr5_-_84565218
Show fit
|
0.54 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5
|
|
chrX_-_7242065
Show fit
|
0.45 |
ENSMUST00000191497.2
ENSMUST00000115744.2
|
Usp27x
|
ubiquitin specific peptidase 27, X chromosome
|
|
chrM_+_9870
Show fit
|
0.45 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L
|
|
chr2_+_87404246
Show fit
|
0.42 |
ENSMUST00000213315.2
ENSMUST00000214773.2
|
Olfr1129
|
olfactory receptor 1129
|
|
chr17_-_37523969
Show fit
|
0.42 |
ENSMUST00000060728.7
ENSMUST00000216318.2
|
Olfr95
|
olfactory receptor 95
|
|
chr16_+_14523696
Show fit
|
0.33 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2
|
|
chrX_+_158491589
Show fit
|
0.33 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr2_-_73284262
Show fit
|
0.32 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr14_-_50586329
Show fit
|
0.31 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735
|
|
chr2_-_45000389
Show fit
|
0.31 |
ENSMUST00000201804.4
ENSMUST00000028229.13
ENSMUST00000202187.4
ENSMUST00000153561.6
ENSMUST00000201490.2
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr7_+_140261864
Show fit
|
0.31 |
ENSMUST00000214637.3
|
Olfr45
|
olfactory receptor 45
|
|
chr1_+_171668122
Show fit
|
0.30 |
ENSMUST00000135386.2
|
Cd84
|
CD84 antigen
|
|
chr3_+_63988968
Show fit
|
0.30 |
ENSMUST00000029406.6
|
Vmn2r1
|
vomeronasal 2, receptor 1
|
|
chr18_+_4920513
Show fit
|
0.30 |
ENSMUST00000126977.8
|
Svil
|
supervillin
|
|
chr15_+_102885467
Show fit
|
0.30 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9
|
|
chr6_-_36787096
Show fit
|
0.30 |
ENSMUST00000201321.2
ENSMUST00000101534.5
|
Ptn
|
pleiotrophin
|
|
chr6_+_57180275
Show fit
|
0.28 |
ENSMUST00000226892.2
ENSMUST00000227421.2
|
Vmn1r13
|
vomeronasal 1 receptor 13
|
|
chr2_+_83554770
Show fit
|
0.28 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V
|
|
chr15_+_31224616
Show fit
|
0.27 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein
|
|
chr13_+_22508759
Show fit
|
0.27 |
ENSMUST00000226225.2
ENSMUST00000227017.2
|
Vmn1r197
|
vomeronasal 1 receptor 197
|
|
chr2_+_71884943
Show fit
|
0.27 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr2_+_87137925
Show fit
|
0.26 |
ENSMUST00000216396.3
|
Olfr1118
|
olfactory receptor 1118
|
|
chr14_+_8283087
Show fit
|
0.26 |
ENSMUST00000206298.3
ENSMUST00000216079.2
|
Olfr720
|
olfactory receptor 720
|
|
chr18_+_7905440
Show fit
|
0.26 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil
|
|
chr7_-_73187369
Show fit
|
0.25 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2
|
|
chr12_-_76224025
Show fit
|
0.24 |
ENSMUST00000101291.11
ENSMUST00000218621.2
ENSMUST00000076634.5
|
Esr2
|
estrogen receptor 2 (beta)
|
|
chr7_+_107497109
Show fit
|
0.23 |
ENSMUST00000209670.4
ENSMUST00000216937.3
|
Olfr472
|
olfactory receptor 472
|
|
chrM_-_14061
Show fit
|
0.23 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6
|
|
chr2_+_88217406
Show fit
|
0.23 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178
|
|
chr12_+_108145802
Show fit
|
0.23 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K
|
|
chr18_+_37453427
Show fit
|
0.22 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5
|
|
chr5_-_102217770
Show fit
|
0.22 |
ENSMUST00000053177.14
ENSMUST00000174698.2
|
Wdfy3
|
WD repeat and FYVE domain containing 3
|
|
chr6_-_119940694
Show fit
|
0.22 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1
|
|
chr13_-_23302027
Show fit
|
0.22 |
ENSMUST00000228656.2
|
Vmn1r217
|
vomeronasal 1 receptor 217
|
|
chr11_-_73742280
Show fit
|
0.21 |
ENSMUST00000213365.2
|
Olfr393
|
olfactory receptor 393
|
|
chr13_-_23302396
Show fit
|
0.21 |
ENSMUST00000227110.2
|
Vmn1r217
|
vomeronasal 1 receptor 217
|
|
chr15_-_36165017
Show fit
|
0.21 |
ENSMUST00000058643.4
|
Fbxo43
|
F-box protein 43
|
|
chr9_+_77959206
Show fit
|
0.20 |
ENSMUST00000024104.9
|
Gcm1
|
glial cells missing homolog 1
|
|
chr10_-_129530155
Show fit
|
0.20 |
ENSMUST00000204641.3
|
Olfr803
|
olfactory receptor 803
|
|
chr7_-_139941566
Show fit
|
0.19 |
ENSMUST00000215023.2
ENSMUST00000216027.2
ENSMUST00000210932.3
ENSMUST00000211031.3
|
Olfr60
|
olfactory receptor 60
|
|
chr6_+_136495818
Show fit
|
0.18 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr2_-_32976378
Show fit
|
0.18 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3
|
|
chr10_-_14593935
Show fit
|
0.17 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1
|
|
chr13_+_49761506
Show fit
|
0.17 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin
|
|
chr13_+_49697919
Show fit
|
0.16 |
ENSMUST00000177948.2
ENSMUST00000021820.14
|
Aspn
|
asporin
|
|
chrM_+_2743
Show fit
|
0.16 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1
|
|
chr2_-_45001141
Show fit
|
0.16 |
ENSMUST00000201969.4
ENSMUST00000201623.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr14_+_8282925
Show fit
|
0.15 |
ENSMUST00000217642.2
|
Olfr720
|
olfactory receptor 720
|
|
chr8_-_26609153
Show fit
|
0.14 |
ENSMUST00000037182.14
|
Hook3
|
hook microtubule tethering protein 3
|
|
chr5_-_31684036
Show fit
|
0.14 |
ENSMUST00000202421.2
ENSMUST00000201769.4
ENSMUST00000065388.11
|
Supt7l
|
SPT7-like, STAGA complex gamma subunit
|
|
chr17_-_46638874
Show fit
|
0.13 |
ENSMUST00000047970.14
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10
|
|
chr2_-_45002902
Show fit
|
0.13 |
ENSMUST00000076836.13
ENSMUST00000176732.8
ENSMUST00000200844.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr15_+_80861966
Show fit
|
0.13 |
ENSMUST00000139517.9
ENSMUST00000137255.3
ENSMUST00000137004.2
|
Sgsm3
|
small G protein signaling modulator 3
|
|
chr3_+_144283355
Show fit
|
0.13 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F
|
|
chr4_-_52859227
Show fit
|
0.13 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273
|
|
chr6_-_67468831
Show fit
|
0.13 |
ENSMUST00000118364.2
|
Il23r
|
interleukin 23 receptor
|
|
chr1_-_155293141
Show fit
|
0.12 |
ENSMUST00000111775.8
ENSMUST00000111774.2
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr11_-_79421397
Show fit
|
0.11 |
ENSMUST00000103236.4
ENSMUST00000170799.8
ENSMUST00000170422.4
|
Evi2a
Evi2
|
ecotropic viral integration site 2a
ecotropic viral integration site 2
|
|
chr10_-_33500583
Show fit
|
0.11 |
ENSMUST00000161692.2
ENSMUST00000160299.2
ENSMUST00000019920.13
|
Clvs2
|
clavesin 2
|
|
chr4_+_35152056
Show fit
|
0.11 |
ENSMUST00000058595.7
|
Ifnk
|
interferon kappa
|
|
chr2_+_87436556
Show fit
|
0.11 |
ENSMUST00000213103.3
ENSMUST00000216580.2
|
Olfr1130
|
olfactory receptor 1130
|
|
chr6_+_124281607
Show fit
|
0.11 |
ENSMUST00000032234.5
ENSMUST00000112541.8
|
Cd163
|
CD163 antigen
|
|
chr10_+_127256736
Show fit
|
0.11 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2
|
|
chr15_-_43733389
Show fit
|
0.11 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74
|
|
chr3_-_146200870
Show fit
|
0.10 |
ENSMUST00000093951.3
|
Spata1
|
spermatogenesis associated 1
|
|
chrX_+_149330371
Show fit
|
0.09 |
ENSMUST00000066337.13
ENSMUST00000112715.2
|
Alas2
|
aminolevulinic acid synthase 2, erythroid
|
|
chr6_+_136495784
Show fit
|
0.09 |
ENSMUST00000032335.13
ENSMUST00000185724.7
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr4_-_24851086
Show fit
|
0.09 |
ENSMUST00000084781.6
ENSMUST00000108218.10
|
Klhl32
|
kelch-like 32
|
|
chr14_-_36641270
Show fit
|
0.09 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2
|
|
chr1_+_33947250
Show fit
|
0.09 |
ENSMUST00000183034.5
|
Dst
|
dystonin
|
|
chr1_-_156766381
Show fit
|
0.08 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr2_-_88519531
Show fit
|
0.08 |
ENSMUST00000213545.2
|
Olfr1195
|
olfactory receptor 1195
|
|
chr6_+_68233361
Show fit
|
0.08 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111
|
|
chr6_+_149226891
Show fit
|
0.08 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1
|
|
chr2_-_86857424
Show fit
|
0.08 |
ENSMUST00000214857.2
ENSMUST00000215972.2
|
Olfr1104
|
olfactory receptor 1104
|
|
chr10_+_127256993
Show fit
|
0.08 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2
|
|
chr17_-_21006419
Show fit
|
0.08 |
ENSMUST00000233605.2
ENSMUST00000232812.2
ENSMUST00000233186.2
ENSMUST00000233164.2
|
Vmn1r228
|
vomeronasal 1 receptor 228
|
|
chrX_-_142716200
Show fit
|
0.07 |
ENSMUST00000112851.8
ENSMUST00000112856.3
ENSMUST00000033642.10
|
Dcx
|
doublecortin
|
|
chr5_-_53864874
Show fit
|
0.07 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor
|
|
chr6_-_87510200
Show fit
|
0.07 |
ENSMUST00000113637.9
ENSMUST00000071024.7
|
Arhgap25
|
Rho GTPase activating protein 25
|
|
chr17_-_29226700
Show fit
|
0.07 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38
|
|
chr7_-_120673761
Show fit
|
0.07 |
ENSMUST00000047194.4
|
Igsf6
|
immunoglobulin superfamily, member 6
|
|
chr7_+_126549692
Show fit
|
0.06 |
ENSMUST00000106335.8
ENSMUST00000146017.3
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr1_+_174192688
Show fit
|
0.06 |
ENSMUST00000217962.2
ENSMUST00000220394.2
|
Olfr417
|
olfactory receptor 417
|
|
chr14_+_50360643
Show fit
|
0.06 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727
|
|
chr7_-_73191484
Show fit
|
0.06 |
ENSMUST00000197642.2
ENSMUST00000026895.14
ENSMUST00000169922.9
|
Chd2
|
chromodomain helicase DNA binding protein 2
|
|
chr6_+_29361408
Show fit
|
0.06 |
ENSMUST00000156163.2
|
Calu
|
calumenin
|
|
chr15_+_38869415
Show fit
|
0.06 |
ENSMUST00000179165.9
|
Fzd6
|
frizzled class receptor 6
|
|
chr7_-_101494472
Show fit
|
0.06 |
ENSMUST00000211566.2
ENSMUST00000094141.7
ENSMUST00000209329.2
|
Folr2
|
folate receptor 2 (fetal)
|
|
chr1_-_133849131
Show fit
|
0.05 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat
|
|
chr13_+_118851214
Show fit
|
0.05 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10
|
|
chr7_+_122723365
Show fit
|
0.05 |
ENSMUST00000205514.2
ENSMUST00000094053.7
|
Tnrc6a
|
trinucleotide repeat containing 6a
|
|
chr7_-_45570828
Show fit
|
0.05 |
ENSMUST00000038876.13
|
Emp3
|
epithelial membrane protein 3
|
|
chr5_+_88073438
Show fit
|
0.05 |
ENSMUST00000001667.13
ENSMUST00000113267.8
|
Csn3
|
casein kappa
|
|
chr9_-_107648144
Show fit
|
0.05 |
ENSMUST00000183248.3
ENSMUST00000182022.8
ENSMUST00000035199.13
ENSMUST00000182659.8
|
Rbm5
|
RNA binding motif protein 5
|
|
chr3_-_68911807
Show fit
|
0.05 |
ENSMUST00000154741.8
ENSMUST00000148031.2
|
Ift80
|
intraflagellar transport 80
|
|
chr7_-_10011933
Show fit
|
0.05 |
ENSMUST00000227719.2
ENSMUST00000228622.2
ENSMUST00000228086.2
|
Vmn1r66
|
vomeronasal 1 receptor 66
|
|
chr4_-_43710231
Show fit
|
0.05 |
ENSMUST00000217544.2
ENSMUST00000107862.3
|
Olfr71
|
olfactory receptor 71
|
|
chr1_+_45834645
Show fit
|
0.05 |
ENSMUST00000147308.2
|
Wdr75
|
WD repeat domain 75
|
|
chr1_-_132318039
Show fit
|
0.04 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2
|
|
chr4_+_19605451
Show fit
|
0.04 |
ENSMUST00000108250.3
|
Gm12353
|
predicted gene 12353
|
|
chr1_+_128079543
Show fit
|
0.04 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1
|
|
chr17_+_17622934
Show fit
|
0.04 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1
|
|
chr2_-_63014622
Show fit
|
0.04 |
ENSMUST00000075052.10
ENSMUST00000112454.8
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7
|
|
chr15_+_58004831
Show fit
|
0.03 |
ENSMUST00000226889.2
|
Wdyhv1
|
WDYHV motif containing 1
|
|
chr17_-_90395771
Show fit
|
0.03 |
ENSMUST00000197268.5
ENSMUST00000173917.8
|
Nrxn1
|
neurexin I
|
|
chr9_+_74883377
Show fit
|
0.03 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A
|
|
chr4_-_129015493
Show fit
|
0.03 |
ENSMUST00000135763.2
ENSMUST00000149763.3
ENSMUST00000164649.8
|
Hpca
|
hippocalcin
|
|
chr6_+_89860642
Show fit
|
0.03 |
ENSMUST00000226345.2
ENSMUST00000204656.3
ENSMUST00000227456.2
ENSMUST00000227047.2
ENSMUST00000226120.2
ENSMUST00000227888.2
ENSMUST00000228183.2
ENSMUST00000227625.2
|
Vmn1r44
|
vomeronasal 1 receptor 44
|
|
chr7_+_140064129
Show fit
|
0.03 |
ENSMUST00000214296.2
ENSMUST00000213715.3
ENSMUST00000217235.2
|
Olfr535
|
olfactory receptor 535
|
|
chr2_-_59955995
Show fit
|
0.02 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr6_+_37847721
Show fit
|
0.02 |
ENSMUST00000031859.14
ENSMUST00000120428.8
|
Trim24
|
tripartite motif-containing 24
|
|
chr3_+_20011201
Show fit
|
0.02 |
ENSMUST00000091309.12
ENSMUST00000108329.8
ENSMUST00000003714.13
|
Cp
|
ceruloplasmin
|
|
chr18_+_34973605
Show fit
|
0.02 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2
|
|
chr10_+_129601351
Show fit
|
0.02 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808
|
|
chr7_-_115423934
Show fit
|
0.02 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6
|
|
chr12_-_113896002
Show fit
|
0.02 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1
|
|
chr1_-_14380418
Show fit
|
0.02 |
ENSMUST00000027066.13
ENSMUST00000168081.9
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr18_+_9707595
Show fit
|
0.02 |
ENSMUST00000234965.2
|
Colec12
|
collectin sub-family member 12
|
|
chr7_-_104250951
Show fit
|
0.02 |
ENSMUST00000216750.2
ENSMUST00000215538.2
|
Olfr655
|
olfactory receptor 655
|
|
chr6_+_34722887
Show fit
|
0.02 |
ENSMUST00000123823.8
ENSMUST00000136907.8
|
Cald1
|
caldesmon 1
|
|
chr2_+_76480606
Show fit
|
0.01 |
ENSMUST00000099986.3
|
Pjvk
|
pejvakin
|
|
chr19_+_44980565
Show fit
|
0.01 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr8_+_26648169
Show fit
|
0.01 |
ENSMUST00000036807.13
ENSMUST00000130231.2
|
Thap1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr1_-_14380327
Show fit
|
0.01 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr1_-_156766351
Show fit
|
0.01 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr10_+_128139191
Show fit
|
0.01 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit
|
|
chr15_+_6673167
Show fit
|
0.01 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein
|
|
chr10_+_101994841
Show fit
|
0.01 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C
|
|
chr6_+_83985684
Show fit
|
0.01 |
ENSMUST00000203803.3
ENSMUST00000204591.3
ENSMUST00000113823.8
ENSMUST00000153860.4
|
Dysf
|
dysferlin
|
|
chr11_-_66059330
Show fit
|
0.01 |
ENSMUST00000080665.10
|
Dnah9
|
dynein, axonemal, heavy chain 9
|
|
chr14_-_56137697
Show fit
|
0.01 |
ENSMUST00000111325.5
|
Sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr6_-_39702381
Show fit
|
0.01 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene
|
|
chr11_+_96183294
Show fit
|
0.01 |
ENSMUST00000173432.3
|
Hoxb6
|
homeobox B6
|
|
chr5_-_72325482
Show fit
|
0.01 |
ENSMUST00000196241.2
ENSMUST00000013693.11
|
Commd8
|
COMM domain containing 8
|
|
chr19_-_13074935
Show fit
|
0.01 |
ENSMUST00000214561.3
|
Olfr1457
|
olfactory receptor 1457
|
|
chr10_-_109669053
Show fit
|
0.01 |
ENSMUST00000238286.2
|
Nav3
|
neuron navigator 3
|
|
chr19_-_47680528
Show fit
|
0.01 |
ENSMUST00000026045.14
ENSMUST00000086923.6
|
Col17a1
|
collagen, type XVII, alpha 1
|
|
chr2_-_63014514
Show fit
|
0.01 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7
|
|
chr2_-_164013033
Show fit
|
0.00 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1
|
|
chr13_+_76115824
Show fit
|
0.00 |
ENSMUST00000022081.3
|
Spata9
|
spermatogenesis associated 9
|
|
chr15_-_3333003
Show fit
|
0.00 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152
|
|
chr5_-_121641461
Show fit
|
0.00 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b
|
|
chr4_-_119349760
Show fit
|
0.00 |
ENSMUST00000049994.8
|
Rimkla
|
ribosomal modification protein rimK-like family member A
|
|
chr10_+_78353262
Show fit
|
0.00 |
ENSMUST00000204477.5
|
Olfr1358
|
olfactory receptor 1358
|
|
chr5_+_65505657
Show fit
|
0.00 |
ENSMUST00000031096.11
|
Klb
|
klotho beta
|
|
chr1_-_37955569
Show fit
|
0.00 |
ENSMUST00000078307.7
|
Lyg2
|
lysozyme G-like 2
|
|
chr3_-_126918491
Show fit
|
0.00 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain
|
|
chr15_-_13173736
Show fit
|
0.00 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6
|
|
chr19_-_12897671
Show fit
|
0.00 |
ENSMUST00000054737.3
|
Olfr1448
|
olfactory receptor 1448
|
|
chr4_-_144291704
Show fit
|
0.00 |
ENSMUST00000105748.2
|
Aadacl4fm2
|
AADACL4 family member 2
|