Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
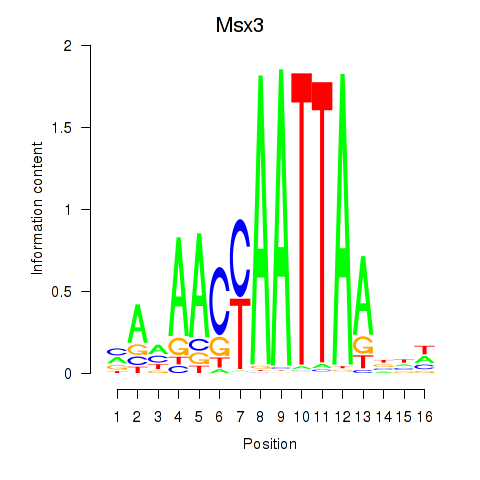
Results for Msx3
Z-value: 0.55
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSMUSG00000025469.11 | msh homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx3 | mm39_v1_chr7_-_139628991_139629007 | 0.57 | 3.1e-01 | Click! |
Activity profile of Msx3 motif
Sorted Z-values of Msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_16485937 | 0.65 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr19_+_13208692 | 0.47 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chrX_-_48980360 | 0.38 |
ENSMUST00000217355.3
|
Olfr1322
|
olfactory receptor 1322 |
| chr7_+_140181182 | 0.36 |
ENSMUST00000214180.2
ENSMUST00000211771.2 |
Olfr46
|
olfactory receptor 46 |
| chr3_-_129834788 | 0.34 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr7_+_3648264 | 0.33 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_-_111820618 | 0.32 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr13_+_23191826 | 0.29 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr2_-_111843053 | 0.29 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr7_-_107633196 | 0.28 |
ENSMUST00000210173.3
|
Olfr478
|
olfactory receptor 478 |
| chr17_-_37523969 | 0.27 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr10_-_75946790 | 0.24 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr2_+_96148418 | 0.20 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr13_+_22508759 | 0.20 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr13_+_23398297 | 0.19 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chr2_+_87696836 | 0.18 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr11_-_73382303 | 0.17 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr16_+_11224481 | 0.16 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr3_+_94745009 | 0.15 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr2_-_45007407 | 0.14 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_32976378 | 0.14 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr3_+_66892979 | 0.14 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr9_+_38516398 | 0.13 |
ENSMUST00000217057.2
|
Olfr914
|
olfactory receptor 914 |
| chr17_+_30940015 | 0.12 |
ENSMUST00000236948.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr5_+_104350475 | 0.11 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr7_-_12829100 | 0.11 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chrX_-_142716200 | 0.11 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr11_-_99134885 | 0.10 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr6_+_68279392 | 0.10 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr3_+_94744844 | 0.10 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr1_+_104696235 | 0.10 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr10_+_85763545 | 0.09 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr2_-_32977182 | 0.09 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr2_+_88348895 | 0.09 |
ENSMUST00000216978.2
ENSMUST00000215912.2 |
Olfr1186
|
olfactory receptor 1186 |
| chr13_+_22268610 | 0.09 |
ENSMUST00000228243.2
ENSMUST00000226680.2 |
Vmn1r188
|
vomeronasal 1 receptor 188 |
| chr5_+_88731386 | 0.09 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr14_+_60615128 | 0.08 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr3_-_97329336 | 0.08 |
ENSMUST00000199297.4
|
Olfr1402
|
olfactory receptor 1402 |
| chr5_+_14564932 | 0.08 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr18_+_4993795 | 0.08 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr12_+_102094977 | 0.07 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr8_+_72973560 | 0.07 |
ENSMUST00000003123.10
|
Fam32a
|
family with sequence similarity 32, member A |
| chr2_-_89671899 | 0.07 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr10_+_39488930 | 0.07 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr10_+_88214989 | 0.07 |
ENSMUST00000127615.8
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr3_+_41697046 | 0.06 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chrX_+_106299484 | 0.06 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr12_-_114443071 | 0.06 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr10_+_88215079 | 0.06 |
ENSMUST00000130301.8
ENSMUST00000020251.10 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr4_-_43710231 | 0.06 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr4_+_134658209 | 0.05 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chrX_+_100492684 | 0.05 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr7_+_84562283 | 0.04 |
ENSMUST00000216367.2
ENSMUST00000214501.3 |
Olfr290
|
olfactory receptor 290 |
| chr17_-_24746804 | 0.04 |
ENSMUST00000176353.8
ENSMUST00000176237.8 |
Traf7
|
TNF receptor-associated factor 7 |
| chr19_+_12257218 | 0.04 |
ENSMUST00000207186.4
ENSMUST00000207915.2 |
Olfr1434
|
olfactory receptor 1434 |
| chr17_-_24746911 | 0.04 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7 |
| chr14_+_122272069 | 0.03 |
ENSMUST00000045976.7
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr1_-_97055931 | 0.03 |
ENSMUST00000027569.14
|
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr3_-_41696906 | 0.03 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr11_+_6366259 | 0.03 |
ENSMUST00000213200.2
|
Ppia
|
peptidylprolyl isomerase A |
| chr15_+_82136598 | 0.02 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr2_+_121787131 | 0.02 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr13_-_23041731 | 0.02 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr4_-_43840201 | 0.02 |
ENSMUST00000214281.2
|
Olfr157
|
olfactory receptor 157 |
| chr9_+_100479732 | 0.02 |
ENSMUST00000124487.8
|
Stag1
|
stromal antigen 1 |
| chr17_+_49922129 | 0.01 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr13_+_22563988 | 0.00 |
ENSMUST00000227685.2
ENSMUST00000227689.2 ENSMUST00000227846.2 |
Vmn1r199
|
vomeronasal 1 receptor 199 |
| chr4_+_102112189 | 0.00 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr7_-_98790275 | 0.00 |
ENSMUST00000037968.10
|
Uvrag
|
UV radiation resistance associated gene |
| chr12_-_113649535 | 0.00 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr9_+_72714156 | 0.00 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chrM_+_9870 | 0.00 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr9_+_96140781 | 0.00 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr6_+_48604157 | 0.00 |
ENSMUST00000203088.3
ENSMUST00000204958.2 |
AI854703
|
expressed sequence AI854703 |
| chr5_-_140986312 | 0.00 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |