Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
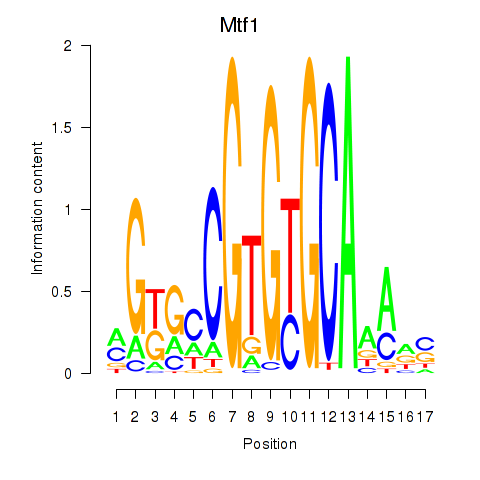
Results for Mtf1
Z-value: 0.65
Transcription factors associated with Mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mtf1
|
ENSMUSG00000028890.14 | metal response element binding transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | mm39_v1_chr4_+_124696336_124696381 | -0.55 | 3.4e-01 | Click! |
Activity profile of Mtf1 motif
Sorted Z-values of Mtf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94905710 | 0.43 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr6_+_18848600 | 0.38 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr15_+_101184488 | 0.35 |
ENSMUST00000229525.2
ENSMUST00000230525.2 |
Atg101
|
autophagy related 101 |
| chr16_+_22676589 | 0.35 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr1_+_127132712 | 0.34 |
ENSMUST00000038361.11
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr11_+_98938137 | 0.32 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr6_+_18848570 | 0.27 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr12_-_36206780 | 0.25 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_36206626 | 0.25 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_36206750 | 0.25 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_+_11972277 | 0.24 |
ENSMUST00000171109.9
ENSMUST00000046948.10 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
| chr5_-_34794185 | 0.22 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr1_-_169359015 | 0.20 |
ENSMUST00000111368.8
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr4_+_133302039 | 0.19 |
ENSMUST00000030662.3
|
Gpatch3
|
G patch domain containing 3 |
| chr9_+_109662063 | 0.18 |
ENSMUST00000197627.5
ENSMUST00000200468.2 |
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr4_+_138700195 | 0.18 |
ENSMUST00000123636.8
ENSMUST00000043042.10 ENSMUST00000050949.9 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr13_-_46118433 | 0.17 |
ENSMUST00000167708.4
ENSMUST00000091628.11 ENSMUST00000180110.9 |
Atxn1
|
ataxin 1 |
| chr8_-_106937555 | 0.17 |
ENSMUST00000035925.7
|
Slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr5_-_34794451 | 0.15 |
ENSMUST00000124668.2
ENSMUST00000001109.11 ENSMUST00000155577.8 ENSMUST00000114329.8 |
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chrX_+_7446721 | 0.15 |
ENSMUST00000115738.8
|
Foxp3
|
forkhead box P3 |
| chr2_+_118943274 | 0.15 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr19_-_41252370 | 0.15 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr19_-_4889314 | 0.14 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr3_+_107803225 | 0.14 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr6_-_83752749 | 0.13 |
ENSMUST00000014892.8
|
Tex261
|
testis expressed gene 261 |
| chr3_+_107803563 | 0.13 |
ENSMUST00000169365.2
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr1_-_169358912 | 0.13 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr2_-_119617985 | 0.13 |
ENSMUST00000110793.8
ENSMUST00000099529.9 ENSMUST00000048493.12 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr14_+_59438658 | 0.13 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr17_-_71833752 | 0.12 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr10_+_20828446 | 0.12 |
ENSMUST00000105525.12
|
Ahi1
|
Abelson helper integration site 1 |
| chr5_+_31026967 | 0.12 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr8_+_106937568 | 0.12 |
ENSMUST00000071592.12
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr7_+_132212349 | 0.12 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr6_-_39095144 | 0.11 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr18_-_77652820 | 0.11 |
ENSMUST00000026494.14
ENSMUST00000182024.2 |
Rnf165
|
ring finger protein 165 |
| chr19_-_4889284 | 0.11 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr1_+_135656885 | 0.11 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr16_+_56942050 | 0.11 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr7_+_43256593 | 0.10 |
ENSMUST00000120935.2
ENSMUST00000127765.8 ENSMUST00000032661.14 |
Zfp819
|
zinc finger protein 819 |
| chr6_-_87827993 | 0.10 |
ENSMUST00000204890.3
ENSMUST00000113617.3 ENSMUST00000113619.8 ENSMUST00000204653.3 ENSMUST00000032138.15 |
Cnbp
|
cellular nucleic acid binding protein |
| chr5_+_57875309 | 0.10 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr6_-_56900917 | 0.10 |
ENSMUST00000031793.8
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr7_-_15656285 | 0.10 |
ENSMUST00000044355.11
|
Selenow
|
selenoprotein W |
| chr11_+_4845328 | 0.09 |
ENSMUST00000038237.8
|
Thoc5
|
THO complex 5 |
| chr17_-_56933872 | 0.09 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr11_+_50022167 | 0.09 |
ENSMUST00000093138.13
ENSMUST00000101270.5 |
Tbc1d9b
|
TBC1 domain family, member 9B |
| chr11_+_4845314 | 0.09 |
ENSMUST00000101615.9
|
Thoc5
|
THO complex 5 |
| chr10_+_77442405 | 0.09 |
ENSMUST00000099538.6
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr8_+_94899292 | 0.08 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr15_-_83033508 | 0.08 |
ENSMUST00000100375.11
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_-_18165876 | 0.08 |
ENSMUST00000125287.9
|
Tango2
|
transport and golgi organization 2 |
| chr5_-_34794546 | 0.08 |
ENSMUST00000114331.10
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chrX_-_8072714 | 0.07 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr15_-_83033559 | 0.07 |
ENSMUST00000058793.14
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr15_+_81686622 | 0.06 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr8_+_124100588 | 0.06 |
ENSMUST00000211932.2
ENSMUST00000212569.2 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr1_-_157240138 | 0.06 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr9_-_14292269 | 0.06 |
ENSMUST00000214236.2
|
Endod1
|
endonuclease domain containing 1 |
| chr14_-_63781381 | 0.06 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr10_-_80837173 | 0.06 |
ENSMUST00000099462.8
ENSMUST00000118233.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr3_-_121325887 | 0.05 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr13_+_24118417 | 0.05 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr16_+_38405718 | 0.05 |
ENSMUST00000165631.2
|
Tmem39a
|
transmembrane protein 39a |
| chr2_-_130506484 | 0.05 |
ENSMUST00000089559.11
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr14_+_59438879 | 0.04 |
ENSMUST00000140136.9
ENSMUST00000142326.2 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr5_+_121798932 | 0.04 |
ENSMUST00000111765.8
|
Brap
|
BRCA1 associated protein |
| chr11_+_4207557 | 0.04 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr8_+_13209141 | 0.04 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr8_+_124100492 | 0.04 |
ENSMUST00000212571.2
ENSMUST00000212470.2 ENSMUST00000108840.4 ENSMUST00000057934.10 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr16_-_22676264 | 0.03 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr19_-_4665509 | 0.03 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_-_46782448 | 0.03 |
ENSMUST00000033142.13
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr7_-_119393182 | 0.03 |
ENSMUST00000106523.8
ENSMUST00000063902.14 ENSMUST00000150844.3 |
Eri2
|
exoribonuclease 2 |
| chr3_+_107803137 | 0.03 |
ENSMUST00000004134.11
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr4_+_134224308 | 0.03 |
ENSMUST00000095074.4
|
Paqr7
|
progestin and adipoQ receptor family member VII |
| chr9_+_108368032 | 0.03 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr19_-_4665668 | 0.03 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_+_66426127 | 0.02 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr3_+_104688363 | 0.02 |
ENSMUST00000002298.7
|
Ppm1j
|
protein phosphatase 1J |
| chr1_+_131794962 | 0.02 |
ENSMUST00000112386.8
ENSMUST00000027693.8 |
Rab29
|
RAB29, member RAS oncogene family |
| chr7_+_119393312 | 0.02 |
ENSMUST00000084644.3
|
Rexo5
|
RNA exonuclease 5 |
| chr6_-_30680502 | 0.02 |
ENSMUST00000133373.8
|
Cep41
|
centrosomal protein 41 |
| chr18_-_9450097 | 0.02 |
ENSMUST00000053917.6
|
Ccny
|
cyclin Y |
| chr15_+_100366886 | 0.02 |
ENSMUST00000037001.10
ENSMUST00000230294.2 |
Letmd1
|
LETM1 domain containing 1 |
| chr15_-_93417380 | 0.01 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr14_+_30673435 | 0.01 |
ENSMUST00000226833.2
|
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr15_+_100366966 | 0.01 |
ENSMUST00000229648.2
|
Letmd1
|
LETM1 domain containing 1 |
| chr2_-_91067212 | 0.01 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr5_-_140986312 | 0.01 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr8_+_93553901 | 0.01 |
ENSMUST00000034187.9
|
Mmp2
|
matrix metallopeptidase 2 |
| chr10_+_29023201 | 0.01 |
ENSMUST00000213243.2
|
Soga3
|
SOGA family member 3 |
| chr1_+_167445815 | 0.01 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr10_+_78749005 | 0.01 |
ENSMUST00000204587.4
ENSMUST00000217073.3 |
Olfr1354
|
olfactory receptor 1354 |
| chr7_+_119393210 | 0.01 |
ENSMUST00000033218.15
ENSMUST00000106520.9 |
Rexo5
|
RNA exonuclease 5 |
| chr6_-_83654789 | 0.01 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr11_+_63023395 | 0.01 |
ENSMUST00000108701.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr11_+_63023893 | 0.01 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr1_+_75376714 | 0.00 |
ENSMUST00000113589.8
|
Speg
|
SPEG complex locus |
| chr6_-_126717590 | 0.00 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr6_-_126717114 | 0.00 |
ENSMUST00000112242.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chrX_-_78715030 | 0.00 |
ENSMUST00000052283.7
|
Mageb16
|
MAGE family member B16 |
| chr18_-_9449947 | 0.00 |
ENSMUST00000234102.2
|
Ccny
|
cyclin Y |
| chr4_-_126057263 | 0.00 |
ENSMUST00000097891.4
|
Sh3d21
|
SH3 domain containing 21 |
| chr10_+_51553852 | 0.00 |
ENSMUST00000122922.10
ENSMUST00000219364.2 |
Rfx6
|
regulatory factor X, 6 |
| chr17_+_71980249 | 0.00 |
ENSMUST00000097284.10
|
Togaram2
|
TOG array regulator of axonemal microtubules 2 |
| chr3_+_55047453 | 0.00 |
ENSMUST00000118963.9
ENSMUST00000061099.14 ENSMUST00000153009.8 |
Ccdc169
|
coiled-coil domain containing 169 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mtf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 0.5 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.2 | GO:0002658 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0072114 | cloaca development(GO:0035844) pronephric nephron tubule development(GO:0039020) pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.3 | GO:0016556 | mRNA modification(GO:0016556) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |