Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
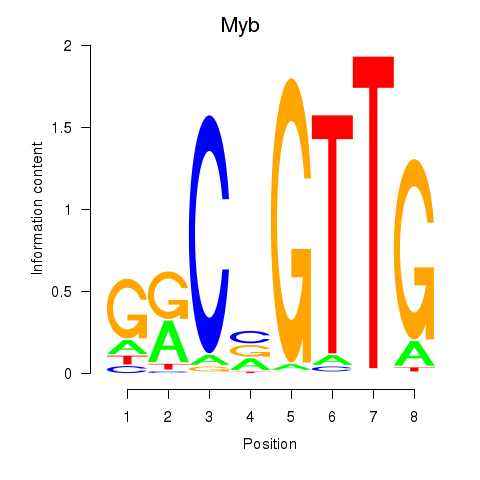
Results for Myb
Z-value: 3.36
Transcription factors associated with Myb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myb
|
ENSMUSG00000019982.16 | myeloblastosis oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myb | mm39_v1_chr10_-_21036792_21036810 | 0.67 | 2.1e-01 | Click! |
Activity profile of Myb motif
Sorted Z-values of Myb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_69812016 | 3.32 |
ENSMUST00000108607.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr3_+_146110387 | 2.23 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr11_+_23615612 | 2.19 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr12_-_90705212 | 1.94 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr8_+_106786190 | 1.69 |
ENSMUST00000109308.3
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr11_-_69811890 | 1.51 |
ENSMUST00000108609.8
ENSMUST00000108608.8 ENSMUST00000164359.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_-_69811717 | 1.49 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr7_+_12758046 | 1.39 |
ENSMUST00000005705.8
|
Trim28
|
tripartite motif-containing 28 |
| chr7_+_46496929 | 1.37 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr11_+_23616007 | 1.32 |
ENSMUST00000058163.11
|
Pus10
|
pseudouridylate synthase 10 |
| chr7_+_46496552 | 1.31 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr11_+_87684548 | 1.30 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr7_+_46496506 | 1.26 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr11_-_118292678 | 1.25 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr14_-_31503869 | 1.22 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr11_+_87684299 | 1.20 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr3_+_88204454 | 1.18 |
ENSMUST00000164166.8
ENSMUST00000168062.8 |
Cct3
|
chaperonin containing Tcp1, subunit 3 (gamma) |
| chr6_-_124410452 | 1.13 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr17_-_80514725 | 1.10 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr11_-_69811347 | 1.09 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr2_+_125994050 | 1.08 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr3_+_88204418 | 1.07 |
ENSMUST00000001452.14
|
Cct3
|
chaperonin containing Tcp1, subunit 3 (gamma) |
| chr19_-_9112919 | 1.05 |
ENSMUST00000049948.6
|
Asrgl1
|
asparaginase like 1 |
| chr7_-_126625739 | 1.05 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_-_137529251 | 1.05 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr12_-_79239022 | 1.04 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr7_-_126101245 | 1.02 |
ENSMUST00000179818.3
|
Atxn2l
|
ataxin 2-like |
| chr16_-_91525485 | 1.01 |
ENSMUST00000231499.2
ENSMUST00000141664.9 ENSMUST00000123751.2 ENSMUST00000122254.8 ENSMUST00000114023.3 |
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr16_+_32698470 | 1.01 |
ENSMUST00000232272.2
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr11_-_74614654 | 1.01 |
ENSMUST00000102520.9
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr16_-_91525655 | 1.01 |
ENSMUST00000117644.8
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr7_-_16657825 | 1.00 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr9_+_106158549 | 1.00 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr10_+_127894816 | 0.97 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr16_-_91525863 | 0.96 |
ENSMUST00000073466.13
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr7_-_45083688 | 0.96 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr14_-_101846459 | 0.95 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr4_+_148025316 | 0.95 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr5_-_31377847 | 0.94 |
ENSMUST00000202294.4
ENSMUST00000031032.11 |
Ppm1g
|
protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform |
| chr14_-_66315144 | 0.94 |
ENSMUST00000022618.6
|
Adam2
|
a disintegrin and metallopeptidase domain 2 |
| chr3_+_137570334 | 0.92 |
ENSMUST00000174561.8
ENSMUST00000173790.8 |
H2az1
|
H2A.Z variant histone 1 |
| chr3_+_103821413 | 0.91 |
ENSMUST00000051139.13
ENSMUST00000068879.11 |
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr1_-_44141574 | 0.89 |
ENSMUST00000143327.2
ENSMUST00000133677.8 ENSMUST00000129702.2 ENSMUST00000149502.8 ENSMUST00000156392.8 ENSMUST00000150911.8 |
Tex30
|
testis expressed 30 |
| chr7_-_126399778 | 0.88 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr2_+_30697838 | 0.88 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr1_+_75119809 | 0.86 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr4_+_128887017 | 0.85 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr8_-_23083751 | 0.85 |
ENSMUST00000009036.11
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr2_-_103627937 | 0.84 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chr2_-_127673738 | 0.84 |
ENSMUST00000028858.8
|
Bub1
|
BUB1, mitotic checkpoint serine/threonine kinase |
| chr7_-_126625657 | 0.83 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_25436884 | 0.82 |
ENSMUST00000114234.2
|
Traf2
|
TNF receptor-associated factor 2 |
| chrX_-_135642025 | 0.81 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr16_+_22926504 | 0.81 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr3_+_103875574 | 0.80 |
ENSMUST00000063717.14
ENSMUST00000055425.15 ENSMUST00000123611.8 ENSMUST00000090685.11 |
Phtf1
|
putative homeodomain transcription factor 1 |
| chr11_+_55360502 | 0.80 |
ENSMUST00000018727.4
|
G3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr2_-_103627334 | 0.80 |
ENSMUST00000111147.8
|
Caprin1
|
cell cycle associated protein 1 |
| chr1_-_44141503 | 0.79 |
ENSMUST00000128190.8
ENSMUST00000147571.8 ENSMUST00000027215.12 ENSMUST00000147661.8 |
Tex30
|
testis expressed 30 |
| chr5_+_123887759 | 0.78 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr11_+_53660834 | 0.78 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chr5_-_137529465 | 0.77 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr8_+_85696453 | 0.77 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr11_+_29642937 | 0.75 |
ENSMUST00000102843.10
ENSMUST00000102842.10 ENSMUST00000078830.11 ENSMUST00000170731.8 |
Rtn4
|
reticulon 4 |
| chr16_+_64672334 | 0.73 |
ENSMUST00000067744.8
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr15_+_101982208 | 0.72 |
ENSMUST00000169681.3
ENSMUST00000229400.2 |
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr4_+_152093260 | 0.72 |
ENSMUST00000097773.4
|
Klhl21
|
kelch-like 21 |
| chr3_-_88857578 | 0.72 |
ENSMUST00000174402.8
ENSMUST00000174077.8 |
Dap3
|
death associated protein 3 |
| chr4_-_118294521 | 0.71 |
ENSMUST00000006565.13
|
Cdc20
|
cell division cycle 20 |
| chr8_+_84442133 | 0.71 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr11_+_22940599 | 0.71 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chrX_-_135641869 | 0.70 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr5_-_122510292 | 0.70 |
ENSMUST00000031419.6
|
Fam216a
|
family with sequence similarity 216, member A |
| chr13_+_119565118 | 0.70 |
ENSMUST00000109203.9
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr4_+_40722911 | 0.70 |
ENSMUST00000164233.8
ENSMUST00000137246.8 ENSMUST00000125442.8 |
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr16_+_32698149 | 0.70 |
ENSMUST00000023489.11
ENSMUST00000171325.9 |
Fyttd1
|
forty-two-three domain containing 1 |
| chr8_-_94738748 | 0.69 |
ENSMUST00000143265.2
|
Amfr
|
autocrine motility factor receptor |
| chr8_-_23083829 | 0.68 |
ENSMUST00000179233.2
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr10_+_79715448 | 0.68 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3 |
| chr2_-_25436952 | 0.67 |
ENSMUST00000028311.13
|
Traf2
|
TNF receptor-associated factor 2 |
| chr2_-_164876690 | 0.67 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr14_+_8508484 | 0.67 |
ENSMUST00000065865.10
ENSMUST00000225891.2 |
Thoc7
|
THO complex 7 |
| chr11_+_116744578 | 0.67 |
ENSMUST00000021173.14
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr17_-_34109513 | 0.66 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr7_-_126625617 | 0.66 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr13_+_51799268 | 0.65 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr5_+_76331727 | 0.65 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr6_+_95094721 | 0.65 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr1_+_179495767 | 0.64 |
ENSMUST00000040538.10
|
Sccpdh
|
saccharopine dehydrogenase (putative) |
| chr8_+_85696396 | 0.64 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chrX_+_74557905 | 0.64 |
ENSMUST00000114070.10
ENSMUST00000033540.6 |
Vbp1
|
von Hippel-Lindau binding protein 1 |
| chr12_+_76812301 | 0.63 |
ENSMUST00000041262.14
ENSMUST00000126408.2 ENSMUST00000110399.3 ENSMUST00000137826.8 |
Churc1
Fntb
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta |
| chr2_-_34803988 | 0.63 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr3_-_90416757 | 0.62 |
ENSMUST00000107343.8
ENSMUST00000001043.14 ENSMUST00000107344.8 ENSMUST00000076639.11 ENSMUST00000107346.8 ENSMUST00000146740.8 ENSMUST00000107342.2 ENSMUST00000049937.13 |
Chtop
|
chromatin target of PRMT1 |
| chr7_-_44465998 | 0.62 |
ENSMUST00000209072.2
ENSMUST00000047356.11 |
Atf5
|
activating transcription factor 5 |
| chr8_-_95564881 | 0.62 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chrX_+_55500170 | 0.62 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr10_+_127894843 | 0.62 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr5_+_129970882 | 0.61 |
ENSMUST00000201855.2
ENSMUST00000073945.6 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr14_-_70391260 | 0.61 |
ENSMUST00000035612.7
|
Ccar2
|
cell cycle activator and apoptosis regulator 2 |
| chr10_+_79951613 | 0.60 |
ENSMUST00000003152.14
|
Stk11
|
serine/threonine kinase 11 |
| chr5_-_121523670 | 0.60 |
ENSMUST00000146185.2
ENSMUST00000042312.14 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr2_+_30306116 | 0.59 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr10_-_22607817 | 0.59 |
ENSMUST00000095794.4
|
Tbpl1
|
TATA box binding protein-like 1 |
| chr1_+_135060994 | 0.59 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr19_-_41373526 | 0.58 |
ENSMUST00000059672.9
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr3_+_137570248 | 0.58 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1 |
| chr14_-_47059546 | 0.58 |
ENSMUST00000226937.2
|
Gmfb
|
glia maturation factor, beta |
| chr13_+_119565424 | 0.58 |
ENSMUST00000026520.14
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr19_+_3901797 | 0.57 |
ENSMUST00000072055.13
|
Chka
|
choline kinase alpha |
| chr10_-_95399997 | 0.57 |
ENSMUST00000020217.7
|
Nudt4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr8_+_84441806 | 0.56 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr2_+_157120946 | 0.56 |
ENSMUST00000116380.9
ENSMUST00000029171.6 |
Rpn2
|
ribophorin II |
| chr13_-_43634695 | 0.56 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr8_-_13155431 | 0.56 |
ENSMUST00000164416.8
|
Pcid2
|
PCI domain containing 2 |
| chr6_-_72416531 | 0.55 |
ENSMUST00000205335.2
ENSMUST00000206692.2 ENSMUST00000059472.10 |
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr6_-_47571901 | 0.55 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr9_-_108444561 | 0.54 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr11_-_79853200 | 0.54 |
ENSMUST00000108241.8
ENSMUST00000043152.6 |
Utp6
|
UTP6 small subunit processome component |
| chr8_-_111758343 | 0.53 |
ENSMUST00000065784.6
|
Ddx19b
|
DEAD box helicase 19b |
| chr16_-_56533179 | 0.53 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr5_-_25047577 | 0.53 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr11_-_101676076 | 0.53 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr17_-_46985181 | 0.53 |
ENSMUST00000024766.7
|
Rrp36
|
ribosomal RNA processing 36 |
| chr18_+_67774659 | 0.52 |
ENSMUST00000025418.4
ENSMUST00000235799.2 |
Psmg2
|
proteasome (prosome, macropain) assembly chaperone 2 |
| chr9_+_44290832 | 0.52 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr5_-_121523634 | 0.52 |
ENSMUST00000120784.8
ENSMUST00000155379.8 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr8_-_85696369 | 0.51 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr8_-_72175949 | 0.51 |
ENSMUST00000125092.2
|
Fcho1
|
FCH domain only 1 |
| chr14_-_61794195 | 0.51 |
ENSMUST00000100496.5
|
Spryd7
|
SPRY domain containing 7 |
| chr4_+_135583018 | 0.51 |
ENSMUST00000105853.10
ENSMUST00000097844.9 ENSMUST00000102544.9 ENSMUST00000126641.2 |
Srsf10
|
serine and arginine-rich splicing factor 10 |
| chr5_+_129970790 | 0.51 |
ENSMUST00000051758.11
|
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr14_+_76714350 | 0.51 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr16_+_17327076 | 0.51 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr19_+_36387123 | 0.51 |
ENSMUST00000225411.2
ENSMUST00000062389.6 |
Pcgf5
|
polycomb group ring finger 5 |
| chr15_-_79718423 | 0.50 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr12_+_102724223 | 0.49 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr6_+_117883732 | 0.49 |
ENSMUST00000179224.8
ENSMUST00000035493.14 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr11_+_22940519 | 0.49 |
ENSMUST00000173867.8
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr14_-_47059694 | 0.49 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chr4_-_129271909 | 0.49 |
ENSMUST00000030610.3
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr17_+_85265420 | 0.48 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr7_-_45084012 | 0.48 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr3_+_146110709 | 0.48 |
ENSMUST00000129978.2
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr15_-_83348735 | 0.48 |
ENSMUST00000229337.2
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr4_-_129636073 | 0.48 |
ENSMUST00000066257.6
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr5_+_33815466 | 0.48 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr10_-_80873138 | 0.47 |
ENSMUST00000059551.7
ENSMUST00000117276.11 |
Slc39a3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr15_-_38518458 | 0.47 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr7_-_18644316 | 0.47 |
ENSMUST00000150065.8
ENSMUST00000098780.10 |
Ccdc61
|
coiled-coil domain containing 61 |
| chr1_-_156546600 | 0.47 |
ENSMUST00000122424.8
ENSMUST00000086153.8 |
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr8_+_88845406 | 0.47 |
ENSMUST00000121097.8
ENSMUST00000117775.2 |
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr11_-_69812053 | 0.46 |
ENSMUST00000108613.10
ENSMUST00000043419.10 ENSMUST00000070996.11 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_75119419 | 0.46 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr4_+_149569717 | 0.46 |
ENSMUST00000030842.8
|
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr9_-_108903117 | 0.45 |
ENSMUST00000161521.8
ENSMUST00000045011.9 |
Atrip
|
ATR interacting protein |
| chr5_+_33978035 | 0.45 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr5_+_135178509 | 0.45 |
ENSMUST00000153183.8
|
Tbl2
|
transducin (beta)-like 2 |
| chr11_-_69871320 | 0.45 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr6_+_122285615 | 0.45 |
ENSMUST00000007602.15
ENSMUST00000112610.2 |
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr11_+_70416185 | 0.45 |
ENSMUST00000018430.7
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr8_-_25215778 | 0.45 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr4_-_119279551 | 0.44 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr11_-_23615862 | 0.44 |
ENSMUST00000020523.4
|
Pex13
|
peroxisomal biogenesis factor 13 |
| chr11_+_105927698 | 0.44 |
ENSMUST00000058438.9
|
Dcaf7
|
DDB1 and CUL4 associated factor 7 |
| chr17_+_28059129 | 0.44 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr17_-_80369626 | 0.44 |
ENSMUST00000184635.8
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr7_+_120450406 | 0.44 |
ENSMUST00000143322.9
ENSMUST00000106488.3 |
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chrX_+_7446721 | 0.44 |
ENSMUST00000115738.8
|
Foxp3
|
forkhead box P3 |
| chr7_-_16121716 | 0.43 |
ENSMUST00000211741.2
ENSMUST00000210999.2 |
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr8_+_88845375 | 0.43 |
ENSMUST00000095214.10
|
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr11_+_120564185 | 0.43 |
ENSMUST00000135346.8
ENSMUST00000127269.8 ENSMUST00000131727.9 ENSMUST00000149389.8 ENSMUST00000153346.8 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr4_-_148711453 | 0.43 |
ENSMUST00000165113.8
ENSMUST00000172073.8 ENSMUST00000105702.9 ENSMUST00000084125.10 |
Tardbp
|
TAR DNA binding protein |
| chr10_+_127126643 | 0.43 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr3_-_88857213 | 0.43 |
ENSMUST00000172942.8
ENSMUST00000107491.11 |
Dap3
|
death associated protein 3 |
| chr6_+_117883783 | 0.43 |
ENSMUST00000177918.8
ENSMUST00000163168.9 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_-_131066004 | 0.43 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr19_-_41252370 | 0.42 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr6_-_30873669 | 0.42 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr17_-_80369762 | 0.42 |
ENSMUST00000061331.14
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr9_-_24414423 | 0.41 |
ENSMUST00000142064.8
ENSMUST00000170356.2 |
Dpy19l1
|
dpy-19-like 1 (C. elegans) |
| chr11_-_70560110 | 0.40 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr5_-_123662175 | 0.40 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr2_+_90677499 | 0.40 |
ENSMUST00000136872.8
ENSMUST00000150232.8 ENSMUST00000111467.4 |
Mtch2
|
mitochondrial carrier 2 |
| chr6_-_124806430 | 0.40 |
ENSMUST00000047510.10
|
Usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr7_+_28937859 | 0.39 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr18_+_46730765 | 0.39 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr1_+_82817170 | 0.39 |
ENSMUST00000189220.7
ENSMUST00000113444.8 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr6_+_97187650 | 0.39 |
ENSMUST00000044681.7
|
Arl6ip5
|
ADP-ribosylation factor-like 6 interacting protein 5 |
| chr5_+_33815910 | 0.39 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr12_+_78273356 | 0.38 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr7_-_100021514 | 0.38 |
ENSMUST00000032963.10
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr2_-_25162347 | 0.38 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr14_+_55909816 | 0.38 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr14_-_25927674 | 0.38 |
ENSMUST00000100811.6
|
Tmem254a
|
transmembrane protein 254a |
| chr7_-_113716996 | 0.38 |
ENSMUST00000069449.7
|
Rras2
|
related RAS viral (r-ras) oncogene 2 |
| chr3_+_14598848 | 0.37 |
ENSMUST00000108370.9
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr6_-_131365380 | 0.37 |
ENSMUST00000032309.13
ENSMUST00000087865.4 |
Ybx3
|
Y box protein 3 |
| chr17_+_71859026 | 0.37 |
ENSMUST00000124001.8
ENSMUST00000167641.8 ENSMUST00000064420.12 |
Spdya
|
speedy/RINGO cell cycle regulator family, member A |
| chr2_+_30306045 | 0.37 |
ENSMUST00000042055.10
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr5_-_121523450 | 0.37 |
ENSMUST00000152265.8
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.8 | 2.5 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.5 | 2.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.4 | 3.9 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 3.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 1.1 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 0.3 | 1.4 | GO:0090309 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) positive regulation of methylation-dependent chromatin silencing(GO:0090309) negative regulation of DNA demethylation(GO:1901536) |
| 0.3 | 1.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 2.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.3 | 1.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.3 | 1.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.2 | 0.7 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.2 | 1.4 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 2.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 3.4 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.9 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.2 | 1.5 | GO:1903719 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.2 | 0.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.2 | 0.5 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 0.7 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.2 | 1.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 0.5 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 | 0.8 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.4 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.4 | GO:0002851 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 1.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.7 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.7 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.4 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.5 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.6 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 1.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.1 | 0.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.4 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 2.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.8 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.6 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.9 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 | 1.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.4 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.5 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.1 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.3 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 2.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.6 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 1.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 2.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.4 | GO:0061484 | negative regulation of mitochondrial membrane potential(GO:0010917) hematopoietic stem cell migration(GO:0035701) hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.9 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 1.1 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:1901228 | positive regulation of skeletal muscle tissue growth(GO:0048633) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:1903903 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.1 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 1.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 1.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.7 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 0.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 1.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.3 | 0.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 0.6 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 3.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 4.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 2.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.4 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.9 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.9 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.3 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0032280 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 1.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.5 | 7.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.5 | 1.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.4 | 1.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 1.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.4 | 1.1 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.3 | 3.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.7 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 0.9 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 3.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 0.7 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 1.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.1 | 0.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.6 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) |
| 0.1 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 2.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.2 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 2.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 3.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 3.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 3.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.8 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 3.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |