Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
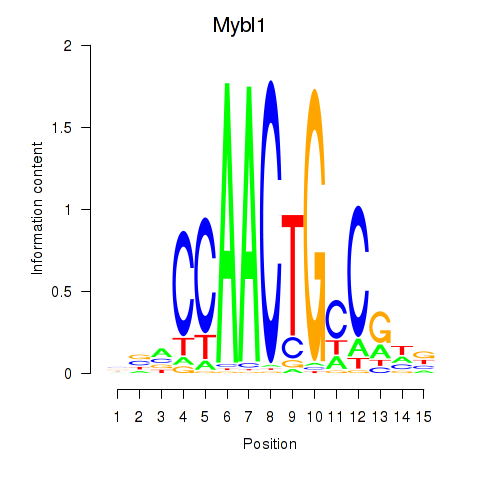
Results for Mybl1
Z-value: 1.33
Transcription factors associated with Mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl1
|
ENSMUSG00000025912.17 | myeloblastosis oncogene-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl1 | mm39_v1_chr1_-_9770434_9770554 | 0.44 | 4.6e-01 | Click! |
Activity profile of Mybl1 motif
Sorted Z-values of Mybl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11618192 | 1.23 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr19_-_11618165 | 1.20 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr2_+_30331839 | 0.81 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr11_+_87684548 | 0.81 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr14_-_56339915 | 0.78 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr11_+_87684299 | 0.77 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr7_-_110443557 | 0.62 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr2_+_150751475 | 0.56 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr17_+_48554786 | 0.55 |
ENSMUST00000048065.6
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr1_-_60137294 | 0.54 |
ENSMUST00000141417.3
ENSMUST00000122038.8 |
Wdr12
|
WD repeat domain 12 |
| chr8_+_95744320 | 0.54 |
ENSMUST00000051259.10
|
Adgrg3
|
adhesion G protein-coupled receptor G3 |
| chr4_+_102617332 | 0.54 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr17_-_34406193 | 0.52 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr12_-_90705212 | 0.52 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr8_+_84442133 | 0.47 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr4_+_150322151 | 0.44 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr9_+_51958453 | 0.43 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr7_+_46496929 | 0.43 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr2_-_105229653 | 0.43 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr1_-_165462020 | 0.41 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr14_-_76248274 | 0.41 |
ENSMUST00000088922.5
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr7_-_119319965 | 0.40 |
ENSMUST00000033236.9
|
Thumpd1
|
THUMP domain containing 1 |
| chr1_-_60137263 | 0.40 |
ENSMUST00000143342.8
|
Wdr12
|
WD repeat domain 12 |
| chr11_+_32233511 | 0.40 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr6_+_85429023 | 0.40 |
ENSMUST00000204592.3
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr17_+_28075415 | 0.40 |
ENSMUST00000114849.3
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr5_+_135038006 | 0.39 |
ENSMUST00000111216.8
ENSMUST00000046999.12 |
Abhd11
|
abhydrolase domain containing 11 |
| chr8_-_111758343 | 0.39 |
ENSMUST00000065784.6
|
Ddx19b
|
DEAD box helicase 19b |
| chr2_-_24864998 | 0.39 |
ENSMUST00000194392.2
|
Mrpl41
|
mitochondrial ribosomal protein L41 |
| chr16_+_22926162 | 0.39 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_+_46496552 | 0.39 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr3_+_88204454 | 0.39 |
ENSMUST00000164166.8
ENSMUST00000168062.8 |
Cct3
|
chaperonin containing Tcp1, subunit 3 (gamma) |
| chr8_-_23083751 | 0.39 |
ENSMUST00000009036.11
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr7_+_16186704 | 0.38 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr9_-_55419442 | 0.38 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr11_+_32246489 | 0.38 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr9_-_109678685 | 0.38 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr7_+_46496506 | 0.37 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr3_+_10077608 | 0.37 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr17_+_87979951 | 0.35 |
ENSMUST00000172855.2
|
Msh2
|
mutS homolog 2 |
| chr8_+_84441806 | 0.35 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr3_+_88204418 | 0.35 |
ENSMUST00000001452.14
|
Cct3
|
chaperonin containing Tcp1, subunit 3 (gamma) |
| chr1_+_91107725 | 0.35 |
ENSMUST00000188475.7
ENSMUST00000097648.6 |
Ramp1
|
receptor (calcitonin) activity modifying protein 1 |
| chr15_-_83439818 | 0.34 |
ENSMUST00000230672.2
ENSMUST00000061882.10 |
Mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr19_-_4978238 | 0.34 |
ENSMUST00000237394.2
ENSMUST00000025851.4 |
Dpp3
|
dipeptidylpeptidase 3 |
| chr10_+_61133549 | 0.34 |
ENSMUST00000219375.2
|
Prf1
|
perforin 1 (pore forming protein) |
| chr12_-_79239022 | 0.34 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr17_+_75024727 | 0.34 |
ENSMUST00000024882.8
ENSMUST00000234751.2 ENSMUST00000234568.2 |
Ttc27
|
tetratricopeptide repeat domain 27 |
| chr2_-_164198427 | 0.33 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr6_-_95695781 | 0.33 |
ENSMUST00000204224.3
|
Suclg2
|
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| chr6_+_89620956 | 0.33 |
ENSMUST00000000828.14
ENSMUST00000101171.3 |
Txnrd3
|
thioredoxin reductase 3 |
| chr10_-_79710468 | 0.32 |
ENSMUST00000092325.11
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr7_-_45083688 | 0.32 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr2_+_19376447 | 0.32 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr2_+_122065230 | 0.32 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr14_+_76714350 | 0.31 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr16_+_20536545 | 0.31 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr13_-_30729242 | 0.31 |
ENSMUST00000042834.4
|
Uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr8_+_75742850 | 0.31 |
ENSMUST00000109940.2
|
Hmgxb4
|
HMG box domain containing 4 |
| chr11_-_100595019 | 0.30 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr15_+_75027089 | 0.30 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr15_+_76784110 | 0.29 |
ENSMUST00000068407.6
ENSMUST00000109793.3 |
Commd5
|
COMM domain containing 5 |
| chr8_-_13155431 | 0.29 |
ENSMUST00000164416.8
|
Pcid2
|
PCI domain containing 2 |
| chr2_+_30697838 | 0.29 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr17_+_34406523 | 0.29 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_101308441 | 0.29 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr7_+_125043806 | 0.29 |
ENSMUST00000033010.9
ENSMUST00000135129.2 |
Kdm8
|
lysine (K)-specific demethylase 8 |
| chr5_-_115257336 | 0.29 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr8_-_23083829 | 0.29 |
ENSMUST00000179233.2
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr1_-_44141574 | 0.29 |
ENSMUST00000143327.2
ENSMUST00000133677.8 ENSMUST00000129702.2 ENSMUST00000149502.8 ENSMUST00000156392.8 ENSMUST00000150911.8 |
Tex30
|
testis expressed 30 |
| chr17_-_85097945 | 0.29 |
ENSMUST00000112308.9
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr11_-_78441584 | 0.28 |
ENSMUST00000103242.5
|
Tmem97
|
transmembrane protein 97 |
| chr13_-_24945844 | 0.28 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr7_+_43093507 | 0.28 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr8_+_85696453 | 0.28 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr14_-_66315144 | 0.28 |
ENSMUST00000022618.6
|
Adam2
|
a disintegrin and metallopeptidase domain 2 |
| chr11_-_68864666 | 0.28 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr5_+_76331727 | 0.28 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr2_+_151385797 | 0.28 |
ENSMUST00000142271.2
|
Fkbp1a
|
FK506 binding protein 1a |
| chr14_+_66043515 | 0.28 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr9_-_106324642 | 0.28 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr8_+_85415935 | 0.28 |
ENSMUST00000125370.10
ENSMUST00000175784.2 |
Trmt1
|
tRNA methyltransferase 1 |
| chr4_+_128887017 | 0.27 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chrX_-_50106844 | 0.27 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr19_-_46917661 | 0.27 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr10_+_59159118 | 0.27 |
ENSMUST00000009789.15
ENSMUST00000092512.11 ENSMUST00000105466.3 |
P4ha1
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
| chr11_-_72441054 | 0.27 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr1_-_65218217 | 0.27 |
ENSMUST00000097709.11
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_+_44290832 | 0.27 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr12_-_110669076 | 0.27 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr13_-_55169100 | 0.27 |
ENSMUST00000148221.8
ENSMUST00000052949.13 |
Hk3
|
hexokinase 3 |
| chrM_+_8603 | 0.27 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr13_+_49835576 | 0.26 |
ENSMUST00000165316.8
ENSMUST00000047363.14 |
Iars
|
isoleucine-tRNA synthetase |
| chr14_+_71011744 | 0.26 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr9_+_65370077 | 0.26 |
ENSMUST00000215170.2
|
Spg21
|
SPG21, maspardin |
| chr14_-_55114989 | 0.26 |
ENSMUST00000168622.2
ENSMUST00000177403.2 |
Ppp1r3e
|
protein phosphatase 1, regulatory subunit 3E |
| chr11_+_101215889 | 0.26 |
ENSMUST00000041095.14
ENSMUST00000107264.2 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr1_+_172139934 | 0.26 |
ENSMUST00000039506.15
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr13_-_24945423 | 0.25 |
ENSMUST00000176890.8
ENSMUST00000175689.8 |
Gmnn
|
geminin |
| chr8_-_23698336 | 0.25 |
ENSMUST00000167004.3
|
Gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr14_+_66043281 | 0.25 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr12_+_100165694 | 0.25 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1 |
| chr17_+_28059129 | 0.25 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr3_-_68952057 | 0.25 |
ENSMUST00000107802.8
|
Trim59
|
tripartite motif-containing 59 |
| chr8_+_85696396 | 0.25 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr1_-_44141503 | 0.25 |
ENSMUST00000128190.8
ENSMUST00000147571.8 ENSMUST00000027215.12 ENSMUST00000147661.8 |
Tex30
|
testis expressed 30 |
| chr19_-_8691460 | 0.25 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr2_+_125089110 | 0.25 |
ENSMUST00000082122.14
|
Dut
|
deoxyuridine triphosphatase |
| chr17_-_26314461 | 0.25 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr2_+_122461079 | 0.24 |
ENSMUST00000239506.1
|
SPATA5L1
|
spermatosis associated 5-like 1 |
| chr9_+_56344700 | 0.24 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr11_+_116744578 | 0.24 |
ENSMUST00000021173.14
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr11_-_6469494 | 0.24 |
ENSMUST00000134489.2
|
Myo1g
|
myosin IG |
| chr11_-_102588536 | 0.24 |
ENSMUST00000164506.3
ENSMUST00000092569.13 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr11_+_62442502 | 0.24 |
ENSMUST00000136938.2
|
Ubb
|
ubiquitin B |
| chr11_-_103588605 | 0.24 |
ENSMUST00000021329.14
|
Gosr2
|
golgi SNAP receptor complex member 2 |
| chr16_+_20536415 | 0.23 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chrX_+_49930311 | 0.23 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr18_+_77032080 | 0.23 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr8_-_25215778 | 0.23 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr3_+_96552895 | 0.23 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chr17_-_28736483 | 0.23 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr15_+_88635852 | 0.23 |
ENSMUST00000041297.15
|
Zbed4
|
zinc finger, BED type containing 4 |
| chr2_+_140012560 | 0.23 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr19_-_33567708 | 0.23 |
ENSMUST00000112508.9
|
Lipo3
|
lipase, member O3 |
| chr8_-_85696369 | 0.23 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr3_-_88857578 | 0.23 |
ENSMUST00000174402.8
ENSMUST00000174077.8 |
Dap3
|
death associated protein 3 |
| chr5_+_135178509 | 0.22 |
ENSMUST00000153183.8
|
Tbl2
|
transducin (beta)-like 2 |
| chr5_+_67464284 | 0.22 |
ENSMUST00000113676.6
ENSMUST00000162372.8 |
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr4_+_107171547 | 0.22 |
ENSMUST00000075693.12
ENSMUST00000139527.8 |
Yipf1
|
Yip1 domain family, member 1 |
| chr1_+_134110142 | 0.22 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr13_-_63036096 | 0.22 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr15_-_44291699 | 0.22 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr8_+_3617958 | 0.22 |
ENSMUST00000136592.2
|
Rps23rg1
|
ribosomal protein S23, retrogene 1 |
| chr3_+_82962823 | 0.22 |
ENSMUST00000150268.8
ENSMUST00000122128.2 |
Plrg1
|
pleiotropic regulator 1 |
| chr8_-_25581354 | 0.22 |
ENSMUST00000125466.2
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr6_+_18848570 | 0.21 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr2_+_144441817 | 0.21 |
ENSMUST00000028917.7
|
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chrX_-_73397181 | 0.21 |
ENSMUST00000114152.2
ENSMUST00000114153.2 ENSMUST00000015433.4 |
Lage3
|
L antigen family, member 3 |
| chr5_+_115061293 | 0.21 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr9_+_14187597 | 0.21 |
ENSMUST00000208222.2
|
Sesn3
|
sestrin 3 |
| chr13_+_55740948 | 0.21 |
ENSMUST00000109905.5
|
Tmed9
|
transmembrane p24 trafficking protein 9 |
| chr1_-_93980278 | 0.21 |
ENSMUST00000027507.9
|
Pdcd1
|
programmed cell death 1 |
| chr16_+_31241085 | 0.21 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr18_-_57108405 | 0.21 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr8_-_85696040 | 0.21 |
ENSMUST00000214133.2
ENSMUST00000147812.8 |
Gm49661
Rnaseh2a
|
predicted gene, 49661 ribonuclease H2, large subunit |
| chr11_-_115426618 | 0.21 |
ENSMUST00000121185.8
ENSMUST00000117589.8 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr14_-_99231754 | 0.21 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr2_-_25436952 | 0.20 |
ENSMUST00000028311.13
|
Traf2
|
TNF receptor-associated factor 2 |
| chr4_+_107171597 | 0.20 |
ENSMUST00000128284.8
ENSMUST00000124650.2 |
Yipf1
|
Yip1 domain family, member 1 |
| chr11_+_3282424 | 0.20 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr13_+_119565118 | 0.20 |
ENSMUST00000109203.9
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr10_-_121462219 | 0.20 |
ENSMUST00000039810.8
ENSMUST00000218004.2 |
Xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr12_-_80690573 | 0.20 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr3_-_129518723 | 0.20 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr7_-_104114384 | 0.20 |
ENSMUST00000076922.6
|
Trim30a
|
tripartite motif-containing 30A |
| chr11_-_99045894 | 0.20 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr7_-_23998735 | 0.20 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61 |
| chr18_+_37898633 | 0.20 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr3_+_68479578 | 0.20 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr8_+_85696216 | 0.19 |
ENSMUST00000109734.8
ENSMUST00000005292.15 |
Prdx2
|
peroxiredoxin 2 |
| chr5_-_65694483 | 0.19 |
ENSMUST00000149167.2
|
Smim14
|
small integral membrane protein 14 |
| chr3_-_68952030 | 0.19 |
ENSMUST00000136512.3
|
Trim59
|
tripartite motif-containing 59 |
| chr17_-_26051871 | 0.19 |
ENSMUST00000236515.2
ENSMUST00000236361.2 |
Stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr3_-_88857213 | 0.19 |
ENSMUST00000172942.8
ENSMUST00000107491.11 |
Dap3
|
death associated protein 3 |
| chr8_+_84441854 | 0.19 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a |
| chr5_+_45650821 | 0.19 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr11_-_59730654 | 0.19 |
ENSMUST00000019517.10
|
Cops3
|
COP9 signalosome subunit 3 |
| chr10_-_80426125 | 0.19 |
ENSMUST00000187646.2
ENSMUST00000191440.7 ENSMUST00000003436.12 |
Abhd17a
|
abhydrolase domain containing 17A |
| chr5_+_123887759 | 0.19 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr2_-_93292708 | 0.19 |
ENSMUST00000123565.8
|
Cd82
|
CD82 antigen |
| chr5_+_21087107 | 0.19 |
ENSMUST00000115259.3
|
Tmem60
|
transmembrane protein 60 |
| chr11_-_60101235 | 0.19 |
ENSMUST00000144942.2
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr12_-_108859123 | 0.19 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr6_+_97187650 | 0.19 |
ENSMUST00000044681.7
|
Arl6ip5
|
ADP-ribosylation factor-like 6 interacting protein 5 |
| chr7_+_28937859 | 0.19 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr9_+_44290787 | 0.19 |
ENSMUST00000066601.13
|
Hyou1
|
hypoxia up-regulated 1 |
| chr4_-_129436465 | 0.19 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr3_+_54600268 | 0.18 |
ENSMUST00000199652.5
|
Supt20
|
SPT20 SAGA complex component |
| chr14_-_31299275 | 0.18 |
ENSMUST00000112027.9
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr13_-_45155616 | 0.18 |
ENSMUST00000072329.15
|
Dtnbp1
|
dystrobrevin binding protein 1 |
| chr18_+_80250102 | 0.18 |
ENSMUST00000127234.8
|
Gm16286
|
predicted gene 16286 |
| chr17_-_35265702 | 0.18 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr3_-_90416757 | 0.18 |
ENSMUST00000107343.8
ENSMUST00000001043.14 ENSMUST00000107344.8 ENSMUST00000076639.11 ENSMUST00000107346.8 ENSMUST00000146740.8 ENSMUST00000107342.2 ENSMUST00000049937.13 |
Chtop
|
chromatin target of PRMT1 |
| chr5_+_135038267 | 0.18 |
ENSMUST00000201890.2
ENSMUST00000154469.8 |
Abhd11
|
abhydrolase domain containing 11 |
| chr7_-_44465998 | 0.18 |
ENSMUST00000209072.2
ENSMUST00000047356.11 |
Atf5
|
activating transcription factor 5 |
| chr9_+_95739650 | 0.18 |
ENSMUST00000034980.9
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr13_-_22190575 | 0.18 |
ENSMUST00000150547.3
|
Prss16
|
protease, serine 16 (thymus) |
| chr2_+_90677499 | 0.18 |
ENSMUST00000136872.8
ENSMUST00000150232.8 ENSMUST00000111467.4 |
Mtch2
|
mitochondrial carrier 2 |
| chr17_+_56316305 | 0.18 |
ENSMUST00000159340.8
|
Mpnd
|
MPN domain containing |
| chr13_-_55677109 | 0.18 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr18_+_34891941 | 0.18 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chrX_+_135145813 | 0.18 |
ENSMUST00000048687.11
|
Tceal9
|
transcription elongation factor A like 9 |
| chr9_+_21323120 | 0.18 |
ENSMUST00000002902.8
|
Qtrt1
|
queuine tRNA-ribosyltransferase catalytic subunit 1 |
| chr9_-_121825028 | 0.18 |
ENSMUST00000216669.2
ENSMUST00000215084.2 ENSMUST00000214533.2 ENSMUST00000217610.2 ENSMUST00000084743.7 |
Pomgnt2
|
protein O-linked mannose beta 1,4-N-acetylglucosaminyltransferase 2 |
| chr19_+_60744385 | 0.18 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr6_-_72416531 | 0.18 |
ENSMUST00000205335.2
ENSMUST00000206692.2 ENSMUST00000059472.10 |
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr9_-_105008972 | 0.18 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr5_-_134643805 | 0.18 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr17_-_71158052 | 0.18 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.3 | 0.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 1.3 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.5 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 1.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.3 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.3 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.2 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 1.1 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.2 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.5 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.2 | GO:2000412 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.8 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.1 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.3 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.3 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.2 | GO:0006244 | dUMP biosynthetic process(GO:0006226) pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.2 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.4 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.6 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0061646 | positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization(GO:0061646) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.4 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.2 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.2 | GO:0061198 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.3 | GO:0051712 | regulation of killing of cells of other organism(GO:0051709) positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:1903722 | late endosomal microautophagy(GO:0061738) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.0 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.3 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0035087 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) small RNA loading onto RISC(GO:0070922) |
| 0.0 | 0.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.0 | GO:0002652 | regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 0.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.2 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.3 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.4 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.3 | GO:0016419 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.1 | 0.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.1 | 0.3 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.3 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.3 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 1.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |