Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
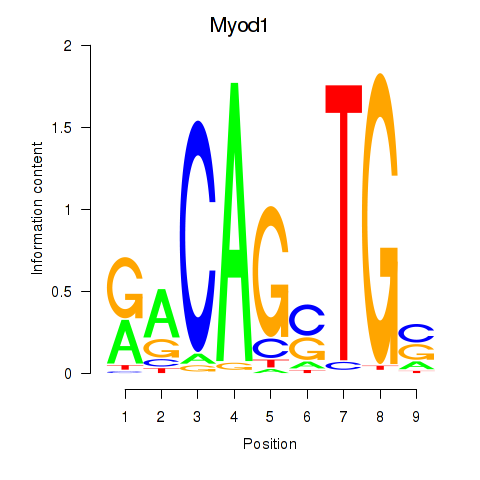
Results for Myod1
Z-value: 0.52
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSMUSG00000009471.5 | myogenic differentiation 1 |
Activity profile of Myod1 motif
Sorted Z-values of Myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_122441719 | 0.42 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr14_-_31503869 | 0.33 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr11_-_96714813 | 0.31 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr17_+_47201552 | 0.27 |
ENSMUST00000040434.9
|
Tbcc
|
tubulin-specific chaperone C |
| chr8_-_34578880 | 0.27 |
ENSMUST00000080152.5
|
Gm10131
|
predicted pseudogene 10131 |
| chr11_-_96714950 | 0.25 |
ENSMUST00000169828.8
ENSMUST00000126949.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr10_+_128745214 | 0.25 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr3_+_41519289 | 0.25 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr11_-_53918916 | 0.25 |
ENSMUST00000020586.7
|
Slc22a4
|
solute carrier family 22 (organic cation transporter), member 4 |
| chr19_+_46345319 | 0.25 |
ENSMUST00000086969.13
|
Mfsd13a
|
major facilitator superfamily domain containing 13a |
| chr3_+_41519355 | 0.24 |
ENSMUST00000191952.2
|
Jade1
|
jade family PHD finger 1 |
| chr19_-_45548942 | 0.24 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr19_+_45549009 | 0.23 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr11_+_87651359 | 0.22 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr4_+_102617332 | 0.22 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_+_133970973 | 0.22 |
ENSMUST00000135228.8
ENSMUST00000144222.8 ENSMUST00000143448.8 ENSMUST00000125921.8 ENSMUST00000122952.8 ENSMUST00000131447.2 |
E130218I03Rik
|
RIKEN cDNA E130218I03 gene |
| chr5_+_28276353 | 0.21 |
ENSMUST00000059155.11
|
Insig1
|
insulin induced gene 1 |
| chr2_+_34661982 | 0.21 |
ENSMUST00000028222.13
ENSMUST00000100171.3 |
Hspa5
|
heat shock protein 5 |
| chr5_+_75735576 | 0.21 |
ENSMUST00000144270.8
ENSMUST00000005815.7 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr4_-_133694607 | 0.21 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr8_+_120121612 | 0.20 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr10_+_126899468 | 0.20 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr7_+_101043568 | 0.19 |
ENSMUST00000098243.4
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_-_131021905 | 0.18 |
ENSMUST00000089510.5
|
Cenpb
|
centromere protein B |
| chr7_-_126625739 | 0.18 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_+_115149170 | 0.18 |
ENSMUST00000031530.9
|
Sppl3
|
signal peptide peptidase 3 |
| chr4_+_106418224 | 0.18 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr9_+_72600721 | 0.17 |
ENSMUST00000238315.2
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4 |
| chr12_+_33004178 | 0.17 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr11_+_101207743 | 0.17 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr7_-_126625657 | 0.17 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_+_103800459 | 0.17 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr10_-_7539009 | 0.16 |
ENSMUST00000163085.8
ENSMUST00000159917.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr7_+_28533279 | 0.16 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr17_+_47816137 | 0.16 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr4_+_148025316 | 0.16 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr14_+_62529924 | 0.16 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr15_+_102379503 | 0.15 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr10_+_61531282 | 0.15 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr12_-_34578842 | 0.15 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr11_-_120538928 | 0.15 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr7_+_141055135 | 0.15 |
ENSMUST00000026585.14
|
Tspan4
|
tetraspanin 4 |
| chr14_-_31552335 | 0.15 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr19_+_4560500 | 0.15 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr10_+_127919142 | 0.15 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr2_+_103800553 | 0.15 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr19_+_46611826 | 0.15 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr8_-_48443525 | 0.14 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr9_+_44308355 | 0.14 |
ENSMUST00000215121.2
ENSMUST00000213388.2 |
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr10_+_75768964 | 0.14 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr9_-_108443916 | 0.14 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr6_+_17749169 | 0.14 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr7_+_141995545 | 0.14 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr3_+_146110387 | 0.14 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr13_-_99027544 | 0.14 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr11_+_109376432 | 0.14 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr9_+_44308149 | 0.14 |
ENSMUST00000215420.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr10_-_7539333 | 0.13 |
ENSMUST00000162606.8
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr3_+_95801325 | 0.13 |
ENSMUST00000197081.2
ENSMUST00000056710.10 |
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr8_+_53964721 | 0.13 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr3_-_19319155 | 0.13 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr7_-_126625617 | 0.13 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr9_+_107852733 | 0.13 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_-_117681864 | 0.13 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr15_-_78657640 | 0.13 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_+_34124078 | 0.13 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr11_+_115705550 | 0.13 |
ENSMUST00000021134.10
ENSMUST00000106481.9 |
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr9_+_107468146 | 0.13 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr2_-_120370333 | 0.13 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr11_+_32405367 | 0.13 |
ENSMUST00000051053.5
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr14_-_30741012 | 0.13 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr9_+_107445101 | 0.13 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr7_-_141934524 | 0.13 |
ENSMUST00000209263.2
|
Gm49369
|
predicted gene, 49369 |
| chr4_+_148225139 | 0.12 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr7_+_121888520 | 0.12 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr1_+_151220222 | 0.12 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr3_+_88523730 | 0.12 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr4_+_118286898 | 0.12 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr11_-_48717482 | 0.12 |
ENSMUST00000104959.2
|
Gm12184
|
predicted gene 12184 |
| chr9_-_108444561 | 0.12 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr3_+_106393348 | 0.12 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr12_-_69243871 | 0.12 |
ENSMUST00000223192.2
|
Gm49383
|
predicted gene, 49383 |
| chr4_-_152122891 | 0.12 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr3_+_51324022 | 0.12 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr13_+_21365308 | 0.12 |
ENSMUST00000221464.2
|
Trim27
|
tripartite motif-containing 27 |
| chr1_+_156386414 | 0.12 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr9_+_120758282 | 0.12 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr4_-_124744454 | 0.12 |
ENSMUST00000125776.8
ENSMUST00000163946.2 ENSMUST00000106190.10 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr7_-_16651107 | 0.12 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr4_-_135000109 | 0.12 |
ENSMUST00000037099.9
|
Clic4
|
chloride intracellular channel 4 (mitochondrial) |
| chr8_-_69636825 | 0.11 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr4_+_129407374 | 0.11 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr15_+_9071761 | 0.11 |
ENSMUST00000189437.8
ENSMUST00000190874.8 |
Nadk2
|
NAD kinase 2, mitochondrial |
| chr17_+_47816074 | 0.11 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr19_+_32463151 | 0.11 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr16_-_95792090 | 0.11 |
ENSMUST00000233128.2
ENSMUST00000023630.16 |
Psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr3_+_40699763 | 0.11 |
ENSMUST00000203353.3
|
Hspa4l
|
heat shock protein 4 like |
| chr12_+_72807985 | 0.10 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr2_+_30306116 | 0.10 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr2_-_101479846 | 0.10 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr9_+_21323120 | 0.10 |
ENSMUST00000002902.8
|
Qtrt1
|
queuine tRNA-ribosyltransferase catalytic subunit 1 |
| chr19_+_60744385 | 0.10 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chrX_+_36059274 | 0.10 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr4_-_133600308 | 0.10 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr3_+_136375839 | 0.10 |
ENSMUST00000070198.14
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr7_+_3697511 | 0.10 |
ENSMUST00000108627.4
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr17_+_53873964 | 0.10 |
ENSMUST00000000724.15
|
Kat2b
|
K(lysine) acetyltransferase 2B |
| chr10_-_75696074 | 0.10 |
ENSMUST00000038169.8
|
Mif
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr5_+_123214332 | 0.10 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr15_-_36792649 | 0.10 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr15_-_103123711 | 0.10 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr17_+_25992761 | 0.10 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr13_-_32522548 | 0.10 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr5_+_35146727 | 0.10 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_-_133694543 | 0.10 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_103857541 | 0.10 |
ENSMUST00000057921.10
ENSMUST00000063347.12 |
Arf2
|
ADP-ribosylation factor 2 |
| chr13_+_52000704 | 0.10 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chrX_-_139857424 | 0.10 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr17_+_47815968 | 0.10 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr4_+_98812144 | 0.10 |
ENSMUST00000169053.2
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr15_+_9071331 | 0.10 |
ENSMUST00000190591.10
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr6_-_148732946 | 0.10 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr11_-_97171294 | 0.09 |
ENSMUST00000167806.8
ENSMUST00000172108.8 ENSMUST00000001480.14 |
Npepps
|
aminopeptidase puromycin sensitive |
| chr9_+_57468217 | 0.09 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr2_-_28730286 | 0.09 |
ENSMUST00000037117.6
ENSMUST00000171404.8 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr10_+_96453408 | 0.09 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr1_-_75195127 | 0.09 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr4_+_149629559 | 0.09 |
ENSMUST00000105692.2
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr9_+_77824646 | 0.09 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr17_+_47816042 | 0.09 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr13_+_83652352 | 0.09 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr15_+_102379621 | 0.09 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr3_+_95496270 | 0.09 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr4_+_129181407 | 0.09 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chrX_-_11994625 | 0.09 |
ENSMUST00000145872.8
|
Bcor
|
BCL6 interacting corepressor |
| chr15_-_89007290 | 0.09 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr13_+_21364330 | 0.09 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr11_+_120123727 | 0.09 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr4_+_148215339 | 0.09 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr6_+_57557978 | 0.09 |
ENSMUST00000031817.10
|
Herc6
|
hect domain and RLD 6 |
| chr2_-_181240921 | 0.08 |
ENSMUST00000060173.9
|
Samd10
|
sterile alpha motif domain containing 10 |
| chr1_-_97904958 | 0.08 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_-_36014892 | 0.08 |
ENSMUST00000097333.10
ENSMUST00000003628.13 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr11_-_115258508 | 0.08 |
ENSMUST00000044152.13
ENSMUST00000106542.9 |
Hid1
|
HID1 domain containing |
| chrX_-_100463810 | 0.08 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr2_-_52225763 | 0.08 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr3_+_68491487 | 0.08 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_+_141055350 | 0.08 |
ENSMUST00000138092.8
ENSMUST00000146305.8 |
Tspan4
|
tetraspanin 4 |
| chr17_+_26780486 | 0.08 |
ENSMUST00000001619.16
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr3_+_89979948 | 0.08 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr13_-_115226666 | 0.08 |
ENSMUST00000109226.5
|
Pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr8_+_123202935 | 0.08 |
ENSMUST00000146634.8
ENSMUST00000134127.2 |
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr5_+_64317550 | 0.08 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr11_-_82761954 | 0.08 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr10_+_126899396 | 0.08 |
ENSMUST00000006911.12
|
Cdk4
|
cyclin-dependent kinase 4 |
| chr4_-_42168603 | 0.08 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr7_+_27307173 | 0.08 |
ENSMUST00000136962.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr4_-_42665763 | 0.08 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr3_-_89152320 | 0.08 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr11_-_102207486 | 0.08 |
ENSMUST00000146896.9
ENSMUST00000079589.11 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr11_-_118306620 | 0.08 |
ENSMUST00000164927.2
|
Cant1
|
calcium activated nucleotidase 1 |
| chrX_-_100463395 | 0.08 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr1_+_163889713 | 0.08 |
ENSMUST00000097491.10
|
Sell
|
selectin, lymphocyte |
| chr2_-_127363251 | 0.08 |
ENSMUST00000028850.15
ENSMUST00000103215.11 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chrX_-_11994589 | 0.08 |
ENSMUST00000123004.2
|
Bcor
|
BCL6 interacting corepressor |
| chr16_-_11072025 | 0.08 |
ENSMUST00000080030.14
|
Gspt1
|
G1 to S phase transition 1 |
| chr11_+_99748741 | 0.08 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr4_+_124744472 | 0.08 |
ENSMUST00000102628.11
|
Yrdc
|
yrdC domain containing (E.coli) |
| chr18_+_32510270 | 0.08 |
ENSMUST00000234857.2
ENSMUST00000234496.2 ENSMUST00000091967.13 ENSMUST00000025239.9 |
Bin1
|
bridging integrator 1 |
| chr15_+_99291455 | 0.08 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr19_+_36387123 | 0.07 |
ENSMUST00000225411.2
ENSMUST00000062389.6 |
Pcgf5
|
polycomb group ring finger 5 |
| chr3_+_65435825 | 0.07 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr9_-_24414423 | 0.07 |
ENSMUST00000142064.8
ENSMUST00000170356.2 |
Dpy19l1
|
dpy-19-like 1 (C. elegans) |
| chr17_+_73144531 | 0.07 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr5_+_35146880 | 0.07 |
ENSMUST00000114285.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_-_116228921 | 0.07 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr1_-_52271455 | 0.07 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr2_-_35322581 | 0.07 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr11_-_84761009 | 0.07 |
ENSMUST00000154915.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr7_-_24771717 | 0.07 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr17_-_36011378 | 0.07 |
ENSMUST00000119825.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr10_-_21036792 | 0.07 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr14_-_55897854 | 0.07 |
ENSMUST00000002400.7
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr5_-_110801213 | 0.07 |
ENSMUST00000042147.6
|
Noc4l
|
NOC4 like |
| chr7_+_24310171 | 0.07 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chrX_+_35592006 | 0.07 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr18_+_69633741 | 0.07 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr7_+_19144950 | 0.07 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr7_+_140796537 | 0.07 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr7_+_27307422 | 0.07 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr7_+_126832399 | 0.07 |
ENSMUST00000056232.7
|
Zfp553
|
zinc finger protein 553 |
| chr4_-_135221926 | 0.07 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr11_-_62172164 | 0.07 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr6_+_39358036 | 0.07 |
ENSMUST00000031986.5
|
Rab19
|
RAB19, member RAS oncogene family |
| chr7_+_126810780 | 0.07 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_+_137570248 | 0.07 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1 |
| chr3_+_40699900 | 0.07 |
ENSMUST00000203496.3
|
Hspa4l
|
heat shock protein 4 like |
| chr16_+_20492267 | 0.07 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr3_+_95801263 | 0.07 |
ENSMUST00000015894.12
|
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr7_-_25089522 | 0.07 |
ENSMUST00000054301.14
|
Lipe
|
lipase, hormone sensitive |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.2 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:1904349 | positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.3 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.0 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.4 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |