Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
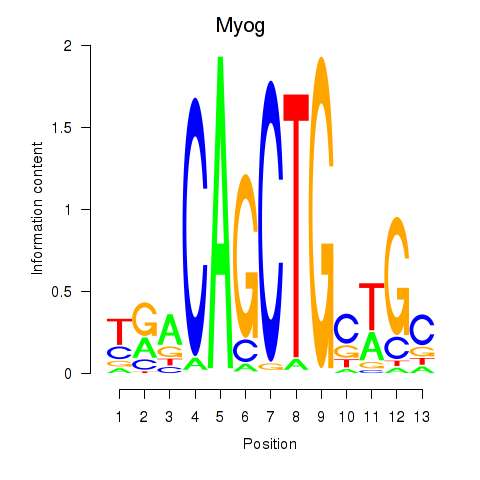
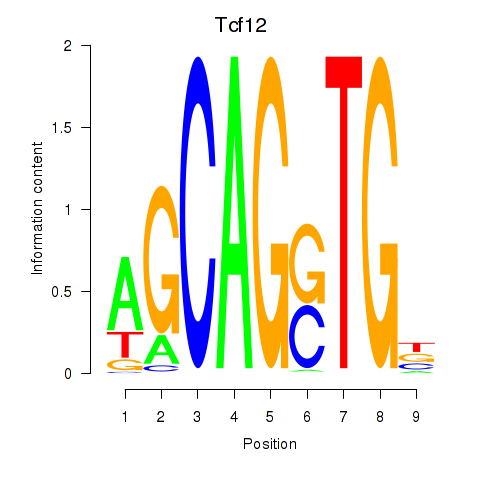
Results for Myog_Tcf12
Z-value: 1.10
Transcription factors associated with Myog_Tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myog
|
ENSMUSG00000026459.6 | myogenin |
|
Tcf12
|
ENSMUSG00000032228.17 | transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf12 | mm39_v1_chr9_-_72018933_72018993 | -0.99 | 1.8e-03 | Click! |
| Myog | mm39_v1_chr1_+_134217727_134217753 | 0.40 | 5.0e-01 | Click! |
Activity profile of Myog_Tcf12 motif
Sorted Z-values of Myog_Tcf12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_96154001 | 1.53 |
ENSMUST00000176059.2
ENSMUST00000177796.2 |
H3c14
|
H3 clustered histone 14 |
| chr3_+_96175970 | 1.50 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr15_-_101268036 | 1.15 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr13_+_23728222 | 1.07 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr7_+_3339059 | 0.97 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339077 | 0.93 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr5_-_137598912 | 0.85 |
ENSMUST00000111007.8
ENSMUST00000133705.2 |
Mospd3
|
motile sperm domain containing 3 |
| chr1_+_177269845 | 0.78 |
ENSMUST00000195002.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr7_-_114162125 | 0.72 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr13_-_21900313 | 0.70 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr7_-_125681577 | 0.67 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
| chr1_-_44258112 | 0.65 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr13_+_23935088 | 0.65 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr2_-_18053158 | 0.64 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chrX_-_72913410 | 0.58 |
ENSMUST00000066576.12
ENSMUST00000114430.8 |
L1cam
|
L1 cell adhesion molecule |
| chr11_+_99755302 | 0.58 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr1_-_160134873 | 0.57 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr12_-_4527138 | 0.56 |
ENSMUST00000085814.5
|
Ncoa1
|
nuclear receptor coactivator 1 |
| chr13_+_47347301 | 0.56 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr10_+_127612243 | 0.56 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chr12_-_45120895 | 0.52 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr13_+_21994588 | 0.50 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chr8_-_123425805 | 0.48 |
ENSMUST00000127984.9
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr1_+_59296065 | 0.48 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr11_+_115790768 | 0.47 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chrX_+_167819606 | 0.47 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr4_+_132968082 | 0.45 |
ENSMUST00000030677.7
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr3_+_135143910 | 0.44 |
ENSMUST00000196446.5
ENSMUST00000106291.10 ENSMUST00000199613.5 |
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_137004793 | 0.44 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr7_+_119495058 | 0.43 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr9_-_48747232 | 0.40 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr10_+_60182630 | 0.39 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr7_-_29204812 | 0.39 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr18_+_61058684 | 0.38 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_+_142025817 | 0.37 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr15_-_58078274 | 0.37 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr13_+_109397184 | 0.37 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr19_+_4282487 | 0.36 |
ENSMUST00000235306.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr5_-_114911509 | 0.36 |
ENSMUST00000086564.11
|
Git2
|
GIT ArfGAP 2 |
| chr4_+_119671688 | 0.36 |
ENSMUST00000106307.9
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr9_-_48747474 | 0.36 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr17_-_47998953 | 0.35 |
ENSMUST00000113301.2
ENSMUST00000113302.10 |
Tomm6
|
translocase of outer mitochondrial membrane 6 |
| chr15_-_77653979 | 0.35 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chrX_+_7439839 | 0.35 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr13_-_21971388 | 0.35 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22 |
| chr2_-_18053595 | 0.34 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr5_-_66308421 | 0.34 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr11_+_69806866 | 0.34 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr11_+_33913013 | 0.33 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr5_+_97145533 | 0.32 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr7_+_142025575 | 0.32 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr13_-_92268156 | 0.32 |
ENSMUST00000151408.9
ENSMUST00000216219.2 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr11_+_77239098 | 0.31 |
ENSMUST00000181283.3
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr9_-_57743989 | 0.31 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr10_+_127595590 | 0.31 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr1_+_87254729 | 0.31 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chrX_-_135769285 | 0.31 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr6_-_83433357 | 0.31 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr8_+_121264161 | 0.31 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr11_+_117700479 | 0.30 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr2_+_32253692 | 0.30 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr2_-_143853122 | 0.30 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr8_+_56747613 | 0.29 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr11_+_77353218 | 0.29 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr3_+_88049875 | 0.29 |
ENSMUST00000107558.9
ENSMUST00000107559.3 |
Mef2d
|
myocyte enhancer factor 2D |
| chr2_-_168432111 | 0.29 |
ENSMUST00000029057.13
ENSMUST00000074618.10 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr1_-_64776890 | 0.29 |
ENSMUST00000116133.4
ENSMUST00000063982.7 |
Fzd5
|
frizzled class receptor 5 |
| chr8_+_66419809 | 0.28 |
ENSMUST00000072482.13
|
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr7_+_27879650 | 0.28 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr17_-_71309012 | 0.28 |
ENSMUST00000128179.2
ENSMUST00000150456.2 ENSMUST00000233357.2 ENSMUST00000233417.2 |
Myl12a
Gm49909
|
myosin, light chain 12A, regulatory, non-sarcomeric predicted gene, 49909 |
| chr4_-_135221810 | 0.28 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr11_+_29323618 | 0.28 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr8_+_106002772 | 0.27 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr10_-_75396164 | 0.27 |
ENSMUST00000051129.10
|
Lrrc75b
|
leucine rich repeat containing 75B |
| chr7_+_3352019 | 0.27 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr5_+_23639432 | 0.27 |
ENSMUST00000094962.9
ENSMUST00000115128.8 |
Kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr16_+_17269845 | 0.27 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr17_+_69071546 | 0.27 |
ENSMUST00000233625.2
|
L3mbtl4
|
L3MBTL4 histone methyl-lysine binding protein |
| chr5_+_21748523 | 0.26 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr2_+_71811526 | 0.26 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_-_125119855 | 0.26 |
ENSMUST00000125053.8
|
Ncor2
|
nuclear receptor co-repressor 2 |
| chr5_-_66308666 | 0.25 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr5_-_21906330 | 0.25 |
ENSMUST00000115217.8
ENSMUST00000060899.9 |
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chrX_-_7242065 | 0.25 |
ENSMUST00000191497.2
ENSMUST00000115744.2 |
Usp27x
|
ubiquitin specific peptidase 27, X chromosome |
| chr7_+_44545501 | 0.25 |
ENSMUST00000071207.14
ENSMUST00000209132.2 ENSMUST00000207069.2 ENSMUST00000209039.2 ENSMUST00000207939.3 ENSMUST00000207485.2 ENSMUST00000208179.2 |
Fuz
|
fuzzy planar cell polarity protein |
| chr16_-_92494203 | 0.25 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr7_+_44465714 | 0.25 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62 |
| chr11_+_116324913 | 0.25 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr3_-_95049655 | 0.25 |
ENSMUST00000013851.4
|
Tnfaip8l2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr11_+_50492899 | 0.25 |
ENSMUST00000142118.3
ENSMUST00000040523.9 |
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 |
| chr5_-_125135434 | 0.24 |
ENSMUST00000134404.6
ENSMUST00000199561.2 |
Ncor2
|
nuclear receptor co-repressor 2 |
| chr2_+_174171979 | 0.24 |
ENSMUST00000109083.2
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_-_170248421 | 0.24 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr5_-_25305621 | 0.24 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_+_10502612 | 0.24 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr2_+_48839505 | 0.24 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr13_-_111945499 | 0.23 |
ENSMUST00000109267.9
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr3_-_20296337 | 0.23 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr2_-_60711706 | 0.23 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_-_52766615 | 0.23 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr7_+_127078371 | 0.23 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr7_+_4122523 | 0.23 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr1_+_59802543 | 0.23 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr2_+_28590159 | 0.23 |
ENSMUST00000074156.7
|
Ak8
|
adenylate kinase 8 |
| chr15_-_83054369 | 0.22 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chrX_+_158410229 | 0.22 |
ENSMUST00000112456.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr13_-_23867924 | 0.22 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr5_-_114911432 | 0.22 |
ENSMUST00000112183.8
|
Git2
|
GIT ArfGAP 2 |
| chr4_-_133856025 | 0.22 |
ENSMUST00000105879.2
ENSMUST00000030651.9 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr2_-_168432235 | 0.22 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr1_+_138891155 | 0.22 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr14_-_55150547 | 0.22 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr9_+_56979307 | 0.22 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr17_+_29077385 | 0.22 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr11_+_83855245 | 0.22 |
ENSMUST00000049714.15
ENSMUST00000092834.12 ENSMUST00000183714.8 ENSMUST00000183456.8 |
Synrg
|
synergin, gamma |
| chr6_+_82379456 | 0.21 |
ENSMUST00000032122.11
|
Tacr1
|
tachykinin receptor 1 |
| chr11_-_94133527 | 0.21 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr17_+_47747657 | 0.21 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr1_+_156386327 | 0.21 |
ENSMUST00000173929.8
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr7_-_24705320 | 0.21 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr10_+_3822667 | 0.21 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_125839897 | 0.21 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr9_-_52079872 | 0.21 |
ENSMUST00000213645.2
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr5_-_114911548 | 0.21 |
ENSMUST00000178440.8
ENSMUST00000043283.14 ENSMUST00000112185.9 ENSMUST00000155908.8 |
Git2
|
GIT ArfGAP 2 |
| chr1_+_128069677 | 0.20 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr19_+_4281953 | 0.20 |
ENSMUST00000025773.5
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr1_+_120048890 | 0.20 |
ENSMUST00000027637.13
ENSMUST00000112644.9 ENSMUST00000056038.15 |
3110009E18Rik
|
RIKEN cDNA 3110009E18 gene |
| chr15_-_103231921 | 0.20 |
ENSMUST00000229551.2
|
Zfp385a
|
zinc finger protein 385A |
| chr6_+_82379768 | 0.20 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr9_-_21963306 | 0.20 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr17_+_34808772 | 0.20 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr7_+_79939747 | 0.20 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr19_-_11796282 | 0.20 |
ENSMUST00000069285.6
|
Stx3
|
syntaxin 3 |
| chr4_-_131802606 | 0.20 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr17_-_23964807 | 0.20 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr12_-_101879375 | 0.20 |
ENSMUST00000177536.8
ENSMUST00000176728.2 ENSMUST00000021605.14 |
Trip11
|
thyroid hormone receptor interactor 11 |
| chr11_+_75400889 | 0.20 |
ENSMUST00000042972.7
|
Rilp
|
Rab interacting lysosomal protein |
| chr5_-_145077048 | 0.20 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr6_+_97783975 | 0.20 |
ENSMUST00000203884.3
ENSMUST00000043637.14 |
Mitf
|
melanogenesis associated transcription factor |
| chr9_-_62888156 | 0.19 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr15_+_61858883 | 0.19 |
ENSMUST00000159338.2
|
Myc
|
myelocytomatosis oncogene |
| chr5_+_117552042 | 0.19 |
ENSMUST00000180430.2
|
Ksr2
|
kinase suppressor of ras 2 |
| chr6_-_85310393 | 0.19 |
ENSMUST00000059034.13
ENSMUST00000045846.12 ENSMUST00000113788.2 |
Sfxn5
|
sideroflexin 5 |
| chr7_-_122969064 | 0.19 |
ENSMUST00000207010.2
ENSMUST00000167309.8 ENSMUST00000205262.2 ENSMUST00000106442.9 ENSMUST00000098060.5 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr5_+_3978032 | 0.19 |
ENSMUST00000143365.8
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr16_+_33504740 | 0.19 |
ENSMUST00000232568.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr8_-_4267260 | 0.19 |
ENSMUST00000168386.9
|
Prr36
|
proline rich 36 |
| chr7_-_140590605 | 0.19 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr5_-_136596299 | 0.19 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr13_-_58758691 | 0.18 |
ENSMUST00000022036.14
|
Slc28a3
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 |
| chr11_+_103061905 | 0.18 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr9_-_29874401 | 0.18 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin |
| chr16_-_4831349 | 0.18 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr8_-_85500998 | 0.18 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr4_-_93223746 | 0.18 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr9_-_43027809 | 0.18 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr7_+_24990596 | 0.18 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr5_-_107873883 | 0.18 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr1_+_143516402 | 0.18 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr6_-_70237939 | 0.18 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr7_-_30251680 | 0.18 |
ENSMUST00000215288.2
ENSMUST00000108165.8 ENSMUST00000153594.3 |
Proser3
|
proline and serine rich 3 |
| chr16_+_51851948 | 0.18 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr2_-_103315483 | 0.18 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr5_-_113968483 | 0.18 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr11_+_6339442 | 0.18 |
ENSMUST00000109786.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr8_+_66070661 | 0.18 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr11_-_100288566 | 0.18 |
ENSMUST00000001592.15
ENSMUST00000107403.2 |
Jup
|
junction plakoglobin |
| chrX_-_73067351 | 0.17 |
ENSMUST00000114353.10
ENSMUST00000101458.9 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chrX_-_12628309 | 0.17 |
ENSMUST00000096495.11
ENSMUST00000076016.6 |
Med14
|
mediator complex subunit 14 |
| chr19_+_10502679 | 0.17 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr15_-_103248512 | 0.17 |
ENSMUST00000168828.3
|
Zfp385a
|
zinc finger protein 385A |
| chr19_+_45036037 | 0.17 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr5_+_150042092 | 0.17 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chrX_+_71006577 | 0.17 |
ENSMUST00000048790.7
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr19_-_40260060 | 0.17 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr2_+_118877610 | 0.17 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr1_-_171910324 | 0.17 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr15_-_66432938 | 0.17 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr9_-_108140925 | 0.17 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr6_-_77956499 | 0.17 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr11_-_98329782 | 0.16 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr16_+_51851917 | 0.16 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr11_+_77384234 | 0.16 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr17_-_34846122 | 0.16 |
ENSMUST00000171376.8
ENSMUST00000169287.2 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr7_+_44146029 | 0.16 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr9_+_108924457 | 0.16 |
ENSMUST00000072093.13
|
Plxnb1
|
plexin B1 |
| chr2_-_5947551 | 0.16 |
ENSMUST00000026924.7
ENSMUST00000095147.9 |
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr7_+_107194446 | 0.16 |
ENSMUST00000040056.15
ENSMUST00000208956.2 |
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr11_+_115054157 | 0.16 |
ENSMUST00000021077.4
|
Slc9a3r1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
| chr7_-_140907471 | 0.16 |
ENSMUST00000211537.2
|
Deaf1
|
DEAF1, transcription factor |
| chr7_-_28001624 | 0.16 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr11_+_68322945 | 0.16 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr11_-_109188917 | 0.16 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr17_-_80787398 | 0.16 |
ENSMUST00000068714.7
|
Sos1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr5_+_72360631 | 0.16 |
ENSMUST00000169617.3
|
Atp10d
|
ATPase, class V, type 10D |
| chr10_+_127637015 | 0.16 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr18_-_74197970 | 0.16 |
ENSMUST00000159162.2
ENSMUST00000091851.10 |
Mapk4
|
mitogen-activated protein kinase 4 |
| chr7_+_44866635 | 0.16 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr12_+_110816687 | 0.16 |
ENSMUST00000221549.2
ENSMUST00000170060.4 |
Zfp839
|
zinc finger protein 839 |
| chr9_+_44893077 | 0.16 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myog_Tcf12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.5 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.5 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.3 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.3 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.4 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.6 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.9 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.3 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.1 | 0.4 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.1 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.1 | 0.3 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.2 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.1 | 0.4 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.3 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0090095 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.4 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.1 | 0.4 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.4 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.1 | GO:0014894 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.2 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.4 | GO:2000828 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.2 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.5 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.5 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.3 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:2000405 | negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.5 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.0 | 0.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.3 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0060397 | development of secondary male sexual characteristics(GO:0046544) JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.4 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0097402 | optic vesicle morphogenesis(GO:0003404) neuroblast migration(GO:0097402) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.1 | GO:0031692 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.4 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0050294 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |