Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
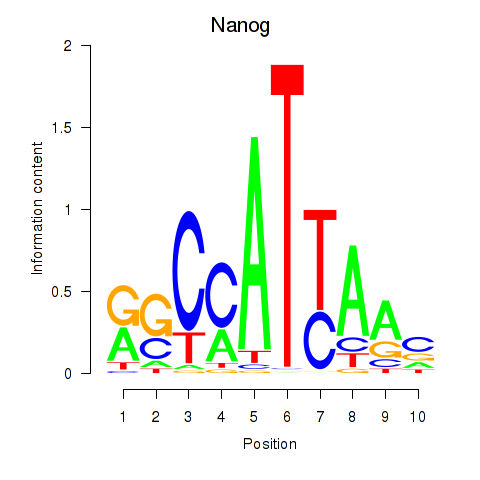
Results for Nanog
Z-value: 0.36
Transcription factors associated with Nanog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nanog
|
ENSMUSG00000012396.13 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nanog | mm39_v1_chr6_+_122684448_122684560 | 0.82 | 9.3e-02 | Click! |
Activity profile of Nanog motif
Sorted Z-values of Nanog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23936250 | 0.81 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr13_-_23945189 | 0.40 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr5_+_107655487 | 0.35 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr3_+_105778174 | 0.24 |
ENSMUST00000164730.2
ENSMUST00000010279.10 |
Adora3
Tmigd3
|
adenosine A3 receptor transmembrane and immunoglobulin domain containing 3 |
| chr4_-_116565509 | 0.23 |
ENSMUST00000030453.5
|
Mmachc
|
methylmalonic aciduria cblC type, with homocystinuria |
| chr7_-_12468931 | 0.23 |
ENSMUST00000233304.2
ENSMUST00000233373.2 ENSMUST00000233874.2 ENSMUST00000233902.2 |
Vmn2r56
|
vomeronasal 2, receptor 56 |
| chr3_-_37366567 | 0.22 |
ENSMUST00000075537.7
ENSMUST00000071400.13 ENSMUST00000102955.11 ENSMUST00000140956.8 |
Cetn4
|
centrin 4 |
| chr11_-_109188917 | 0.22 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr7_-_25454177 | 0.21 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr9_+_39932760 | 0.20 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr10_+_129153986 | 0.19 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr12_+_24701273 | 0.18 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr15_-_71826243 | 0.18 |
ENSMUST00000229585.2
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr19_-_13126896 | 0.18 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459 |
| chr10_+_53213763 | 0.18 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr6_-_116693849 | 0.17 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr7_+_100435548 | 0.16 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr7_+_128290204 | 0.16 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr2_+_155078449 | 0.16 |
ENSMUST00000109682.9
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr5_-_116427003 | 0.16 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr7_-_12342715 | 0.15 |
ENSMUST00000233892.2
|
Vmn2r53
|
vomeronasal 2, receptor 53 |
| chr10_+_129072073 | 0.15 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr7_-_25454126 | 0.15 |
ENSMUST00000108401.3
ENSMUST00000043765.14 |
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr7_+_110368037 | 0.15 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr3_+_105811712 | 0.15 |
ENSMUST00000000574.3
|
Adora3
|
adenosine A3 receptor |
| chr2_-_111820618 | 0.15 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr10_+_78864575 | 0.14 |
ENSMUST00000203906.3
|
Olfr57
|
olfactory receptor 57 |
| chr2_+_3514071 | 0.14 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr15_-_50752437 | 0.14 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr4_+_107825529 | 0.14 |
ENSMUST00000106713.5
ENSMUST00000238795.2 |
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr12_-_72455708 | 0.13 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr13_-_19917092 | 0.13 |
ENSMUST00000151029.3
|
Gpr141b
|
G protein-coupled receptor 141B |
| chr12_+_44268134 | 0.13 |
ENSMUST00000122902.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr2_+_134627987 | 0.12 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chr12_-_115916604 | 0.12 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr2_+_37078210 | 0.12 |
ENSMUST00000213969.2
|
Olfr365
|
olfactory receptor 365 |
| chrX_+_128650486 | 0.12 |
ENSMUST00000167619.9
ENSMUST00000037854.15 |
Diaph2
|
diaphanous related formin 2 |
| chr7_-_141241632 | 0.12 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr7_-_98305737 | 0.12 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr7_+_24967094 | 0.12 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr2_-_45000389 | 0.12 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_59320257 | 0.11 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr18_+_39439778 | 0.11 |
ENSMUST00000235660.2
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr10_-_7162196 | 0.11 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr11_+_68979308 | 0.11 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr7_+_127845984 | 0.11 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr8_-_84996976 | 0.11 |
ENSMUST00000005120.12
ENSMUST00000163993.2 ENSMUST00000098578.10 |
Ccdc130
|
coiled-coil domain containing 130 |
| chr11_+_68979332 | 0.11 |
ENSMUST00000117780.2
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr5_+_135197228 | 0.11 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr7_-_98305986 | 0.11 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr5_-_104125226 | 0.11 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr2_-_86882616 | 0.10 |
ENSMUST00000213781.2
ENSMUST00000217650.2 |
Olfr1106
|
olfactory receptor 1106 |
| chr9_-_70410611 | 0.10 |
ENSMUST00000215848.2
ENSMUST00000113595.2 ENSMUST00000213647.2 |
Rnf111
|
ring finger 111 |
| chr19_-_12100560 | 0.10 |
ENSMUST00000213471.2
ENSMUST00000214676.2 |
Olfr76
|
olfactory receptor 76 |
| chr17_+_38456172 | 0.10 |
ENSMUST00000215078.3
ENSMUST00000215549.3 ENSMUST00000173610.2 |
Olfr133
|
olfactory receptor 133 |
| chr4_+_125918333 | 0.10 |
ENSMUST00000106162.8
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr4_+_150171822 | 0.10 |
ENSMUST00000094451.4
|
Gpr157
|
G protein-coupled receptor 157 |
| chr4_-_116001737 | 0.10 |
ENSMUST00000030469.5
|
Lurap1
|
leucine rich adaptor protein 1 |
| chr6_+_90179768 | 0.10 |
ENSMUST00000078371.6
|
V1ra8
|
vomeronasal 1 receptor, A8 |
| chr10_-_98962187 | 0.10 |
ENSMUST00000060761.7
|
Phxr2
|
per-hexamer repeat gene 2 |
| chr3_-_57599956 | 0.10 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chr16_+_3690232 | 0.09 |
ENSMUST00000151988.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr2_+_28950250 | 0.09 |
ENSMUST00000100237.4
|
Ttf1
|
transcription termination factor, RNA polymerase I |
| chr14_-_78774201 | 0.09 |
ENSMUST00000123853.9
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chrX_+_13527592 | 0.09 |
ENSMUST00000053659.2
|
Gpr82
|
G protein-coupled receptor 82 |
| chr9_+_106245792 | 0.09 |
ENSMUST00000172306.3
|
Dusp7
|
dual specificity phosphatase 7 |
| chr3_+_103767581 | 0.09 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr1_+_153750081 | 0.08 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr17_+_36152383 | 0.08 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr15_-_58828321 | 0.08 |
ENSMUST00000228067.2
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr2_-_166904625 | 0.08 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr8_-_84321032 | 0.08 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr10_+_110581293 | 0.08 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr18_-_35348049 | 0.08 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr17_-_37523969 | 0.08 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr2_+_155078522 | 0.08 |
ENSMUST00000150602.2
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr7_-_30259025 | 0.08 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr3_-_144466602 | 0.08 |
ENSMUST00000059091.6
|
Clca3a1
|
chloride channel accessory 3A1 |
| chr7_+_23330147 | 0.07 |
ENSMUST00000227774.2
ENSMUST00000226771.2 ENSMUST00000228681.2 ENSMUST00000228559.2 ENSMUST00000228674.2 ENSMUST00000227866.2 ENSMUST00000227386.2 ENSMUST00000228484.2 ENSMUST00000226321.2 ENSMUST00000226128.2 ENSMUST00000226733.2 ENSMUST00000228228.2 |
Vmn1r171
|
vomeronasal 1 receptor 171 |
| chr5_+_135197137 | 0.07 |
ENSMUST00000031692.12
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr12_+_108145802 | 0.07 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr19_+_13208692 | 0.07 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr6_-_136899167 | 0.07 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr14_+_51333816 | 0.07 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr5_-_108017019 | 0.07 |
ENSMUST00000128723.8
ENSMUST00000124034.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr2_+_32253692 | 0.07 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr8_+_106024294 | 0.07 |
ENSMUST00000015003.10
|
E2f4
|
E2F transcription factor 4 |
| chr6_+_116627635 | 0.07 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chrX_+_56008685 | 0.07 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chrX_+_7688528 | 0.07 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr1_-_92107971 | 0.06 |
ENSMUST00000186002.3
ENSMUST00000097644.9 |
Hdac4
|
histone deacetylase 4 |
| chr7_+_79309938 | 0.06 |
ENSMUST00000035977.9
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr9_-_15268958 | 0.06 |
ENSMUST00000161132.9
ENSMUST00000098979.11 |
Cep295
|
centrosomal protein 295 |
| chr7_-_30259253 | 0.06 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr11_+_70453806 | 0.06 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr10_+_121477699 | 0.06 |
ENSMUST00000120642.9
ENSMUST00000132744.2 |
D930020B18Rik
|
RIKEN cDNA D930020B18 gene |
| chr17_+_30940015 | 0.06 |
ENSMUST00000236948.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr16_+_10652910 | 0.06 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr2_-_153079828 | 0.06 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr9_-_107512511 | 0.06 |
ENSMUST00000192615.6
ENSMUST00000192837.2 ENSMUST00000193876.2 |
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr5_-_124387812 | 0.06 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr7_+_28580847 | 0.06 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr9_-_107512566 | 0.06 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr7_+_126184108 | 0.06 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr15_+_102898966 | 0.06 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr8_-_84321069 | 0.06 |
ENSMUST00000019382.17
ENSMUST00000212630.2 ENSMUST00000165740.9 |
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr2_+_18069375 | 0.06 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr16_+_18630522 | 0.06 |
ENSMUST00000115578.10
|
Ufd1
|
ubiquitin recognition factor in ER-associated degradation 1 |
| chr11_+_95603494 | 0.06 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr11_+_66802807 | 0.06 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr11_-_114825089 | 0.05 |
ENSMUST00000149663.4
ENSMUST00000239005.2 ENSMUST00000106581.5 |
Cd300lb
|
CD300 molecule like family member B |
| chrX_+_8137881 | 0.05 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_158623460 | 0.05 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chrX_+_101163053 | 0.05 |
ENSMUST00000113627.4
|
Pin4
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr4_+_136038301 | 0.05 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr16_-_20245071 | 0.05 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr9_+_107456086 | 0.05 |
ENSMUST00000149638.8
ENSMUST00000139274.8 ENSMUST00000130053.8 ENSMUST00000122985.8 ENSMUST00000127380.8 ENSMUST00000139581.2 |
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr3_+_79791798 | 0.05 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr17_+_41121979 | 0.05 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr16_-_20245138 | 0.05 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_-_9876815 | 0.05 |
ENSMUST00000237147.2
ENSMUST00000025562.9 |
Incenp
|
inner centromere protein |
| chr9_-_70564403 | 0.05 |
ENSMUST00000213380.2
ENSMUST00000049031.6 |
Mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr19_-_56810593 | 0.05 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr2_-_45001141 | 0.05 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_136038263 | 0.05 |
ENSMUST00000105850.8
ENSMUST00000148843.10 |
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_-_36641456 | 0.05 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr19_+_6096606 | 0.05 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr2_+_115412148 | 0.05 |
ENSMUST00000166472.8
ENSMUST00000110918.3 |
Cdin1
|
CDAN1 interacting nuclease 1 |
| chr17_-_37472385 | 0.05 |
ENSMUST00000219235.3
|
Olfr93
|
olfactory receptor 93 |
| chr10_-_25076008 | 0.05 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr17_-_57554631 | 0.05 |
ENSMUST00000233568.2
ENSMUST00000005975.8 |
Gpr108
|
G protein-coupled receptor 108 |
| chr1_-_171910324 | 0.05 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr10_-_95678748 | 0.04 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr11_-_23845207 | 0.04 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr2_-_45002902 | 0.04 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_19818718 | 0.04 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr1_+_155911136 | 0.04 |
ENSMUST00000111757.10
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr10_-_95678786 | 0.04 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr11_-_99134885 | 0.04 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr6_+_8259288 | 0.04 |
ENSMUST00000159335.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr7_-_26895141 | 0.04 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr2_-_63928339 | 0.04 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr15_-_82872073 | 0.04 |
ENSMUST00000229439.2
|
Tcf20
|
transcription factor 20 |
| chr14_-_20714634 | 0.04 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr2_+_172994841 | 0.04 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr4_-_43710231 | 0.04 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr11_+_58549846 | 0.04 |
ENSMUST00000213232.2
|
Olfr322
|
olfactory receptor 322 |
| chr4_-_46650157 | 0.04 |
ENSMUST00000084621.12
ENSMUST00000107750.2 |
Tbc1d2
|
TBC1 domain family, member 2 |
| chr16_-_3690243 | 0.04 |
ENSMUST00000090522.5
|
Zfp597
|
zinc finger protein 597 |
| chr7_-_102566717 | 0.04 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr6_+_125529911 | 0.04 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr7_-_102368277 | 0.04 |
ENSMUST00000216312.2
|
Olfr33
|
olfactory receptor 33 |
| chr11_+_70861007 | 0.03 |
ENSMUST00000018593.10
|
Rpain
|
RPA interacting protein |
| chr4_+_136038243 | 0.03 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_-_166904902 | 0.03 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr12_+_102094977 | 0.03 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr3_+_60503051 | 0.03 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr13_+_118851214 | 0.03 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr7_-_126183716 | 0.03 |
ENSMUST00000150311.8
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr4_+_43058938 | 0.03 |
ENSMUST00000207569.2
ENSMUST00000079978.13 |
Unc13b
|
unc-13 homolog B |
| chr3_+_122039206 | 0.03 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr11_+_62711295 | 0.03 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr15_+_44320918 | 0.03 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr5_-_100521343 | 0.03 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr9_+_21914083 | 0.03 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr13_+_12580772 | 0.03 |
ENSMUST00000220811.2
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chrX_+_47197789 | 0.03 |
ENSMUST00000114998.8
ENSMUST00000115000.4 |
Xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr10_+_129094996 | 0.03 |
ENSMUST00000213512.2
|
Olfr776
|
olfactory receptor 776 |
| chr19_-_8382424 | 0.03 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr14_-_96756503 | 0.03 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr8_-_37200051 | 0.03 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr19_-_7183626 | 0.03 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr16_-_45544960 | 0.03 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr19_-_7183596 | 0.03 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr18_+_36431732 | 0.03 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr19_+_7534816 | 0.03 |
ENSMUST00000136465.8
ENSMUST00000025925.11 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr11_-_101062111 | 0.03 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr9_-_66421868 | 0.03 |
ENSMUST00000056890.10
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr7_+_63937413 | 0.03 |
ENSMUST00000032736.11
|
Mtmr10
|
myotubularin related protein 10 |
| chr13_-_89890363 | 0.02 |
ENSMUST00000109543.9
ENSMUST00000159337.8 ENSMUST00000159910.8 ENSMUST00000109544.9 |
Vcan
|
versican |
| chr7_-_42962487 | 0.02 |
ENSMUST00000135130.2
ENSMUST00000139061.2 |
Zfp715
|
zinc finger protein 715 |
| chr11_-_121410152 | 0.02 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr2_+_29236815 | 0.02 |
ENSMUST00000028139.11
ENSMUST00000113830.11 |
Med27
|
mediator complex subunit 27 |
| chr16_+_13176238 | 0.02 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr19_+_29499671 | 0.02 |
ENSMUST00000043610.13
|
Ric1
|
RAB6A GEF complex partner 1 |
| chr13_-_3943556 | 0.02 |
ENSMUST00000099946.6
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr11_+_58549642 | 0.02 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr1_-_182109773 | 0.02 |
ENSMUST00000133052.2
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr7_-_28092113 | 0.02 |
ENSMUST00000003536.9
|
Med29
|
mediator complex subunit 29 |
| chr3_-_80710097 | 0.02 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr6_-_102441628 | 0.02 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr1_+_46464625 | 0.02 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C |
| chr18_+_82572595 | 0.02 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr11_-_40624200 | 0.02 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr7_-_85974838 | 0.02 |
ENSMUST00000214977.2
|
Olfr308
|
olfactory receptor 308 |
| chr3_+_103740056 | 0.02 |
ENSMUST00000106822.2
|
Bcl2l15
|
BCLl2-like 15 |
| chr12_+_37930169 | 0.02 |
ENSMUST00000221176.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr3_-_152373997 | 0.02 |
ENSMUST00000045262.11
|
Ak5
|
adenylate kinase 5 |
| chr13_-_3943433 | 0.02 |
ENSMUST00000222504.2
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr15_-_103160082 | 0.02 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nanog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0060466 | interleukin-12-mediated signaling pathway(GO:0035722) activation of meiosis involved in egg activation(GO:0060466) cellular response to interleukin-12(GO:0071349) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.4 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.1 | GO:0014854 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.0 | 0.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |