Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 0.46
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod2
|
ENSMUSG00000038255.7 | neurogenic differentiation 2 |
|
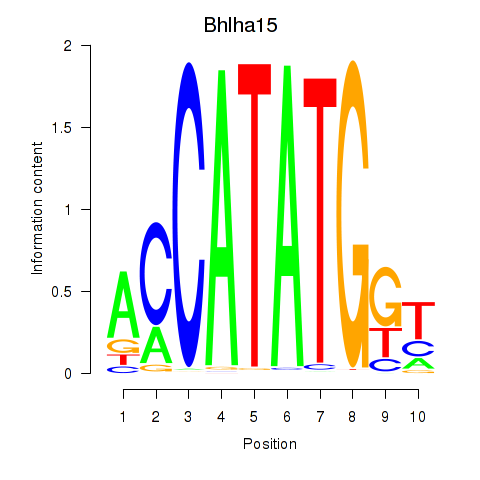
Bhlha15
|
ENSMUSG00000052271.8 | basic helix-loop-helix family, member a15 |
|
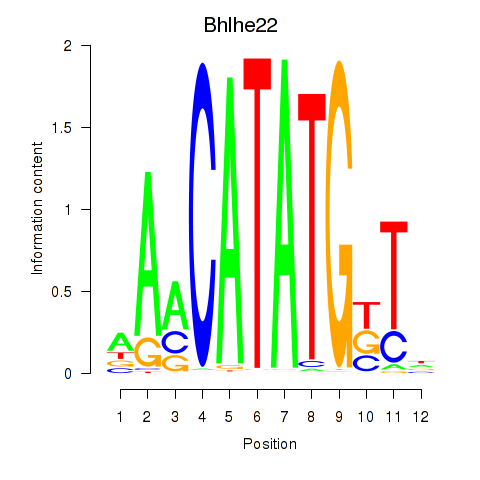
Bhlhe22
|
ENSMUSG00000025128.8 | basic helix-loop-helix family, member e22 |
|
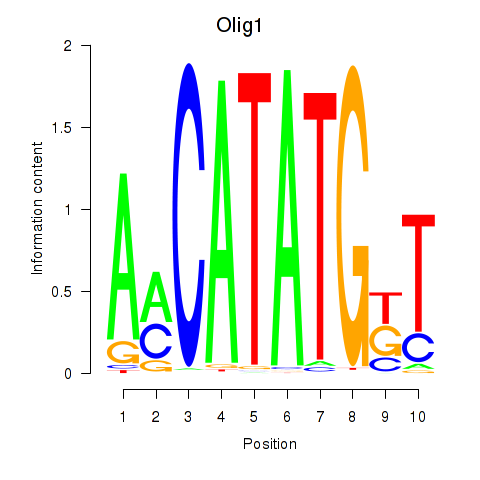
Olig1
|
ENSMUSG00000046160.7 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod2 | mm39_v1_chr11_-_98220466_98220482 | 0.73 | 1.6e-01 | Click! |
| Bhlhe22 | mm39_v1_chr3_+_18108313_18108338 | 0.44 | 4.6e-01 | Click! |
| Bhlha15 | mm39_v1_chr5_+_144127102_144127152 | 0.30 | 6.2e-01 | Click! |
Activity profile of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
Sorted Z-values of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_44258112 | 0.93 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr5_-_104125226 | 0.36 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr3_-_129834788 | 0.35 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr16_-_17540805 | 0.31 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr16_-_17540685 | 0.30 |
ENSMUST00000232163.2
ENSMUST00000232202.2 ENSMUST00000080936.14 ENSMUST00000232645.2 ENSMUST00000232431.2 |
Med15
|
mediator complex subunit 15 |
| chr8_+_26091607 | 0.30 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr10_-_126877382 | 0.30 |
ENSMUST00000116231.4
|
Eef1akmt3
|
EEF1A lysine methyltransferase 3 |
| chr9_+_72892850 | 0.29 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr1_+_150268544 | 0.27 |
ENSMUST00000124973.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr8_+_26092252 | 0.27 |
ENSMUST00000136107.9
ENSMUST00000146919.8 ENSMUST00000139966.8 ENSMUST00000142395.8 ENSMUST00000143445.2 |
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr8_-_5155347 | 0.26 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr6_-_124208815 | 0.24 |
ENSMUST00000233936.2
ENSMUST00000100968.4 |
Vmn2r27
|
vomeronasal 2, receptor27 |
| chr3_-_107240989 | 0.24 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr9_+_19716202 | 0.21 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr8_-_107064615 | 0.19 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr1_+_165591315 | 0.17 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr15_+_99870787 | 0.17 |
ENSMUST00000231160.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr10_+_127595590 | 0.17 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr7_+_11608557 | 0.16 |
ENSMUST00000227611.2
ENSMUST00000226622.2 ENSMUST00000228646.2 ENSMUST00000226855.2 ENSMUST00000228268.2 ENSMUST00000228463.2 |
Vmn1r75
|
vomeronasal 1 receptor 75 |
| chr7_-_23510068 | 0.16 |
ENSMUST00000228383.2
|
Vmn1r175
|
vomeronasal 1 receptor 175 |
| chr11_-_69304501 | 0.15 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr7_+_23330147 | 0.14 |
ENSMUST00000227774.2
ENSMUST00000226771.2 ENSMUST00000228681.2 ENSMUST00000228559.2 ENSMUST00000228674.2 ENSMUST00000227866.2 ENSMUST00000227386.2 ENSMUST00000228484.2 ENSMUST00000226321.2 ENSMUST00000226128.2 ENSMUST00000226733.2 ENSMUST00000228228.2 |
Vmn1r171
|
vomeronasal 1 receptor 171 |
| chr11_+_58845502 | 0.13 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr17_-_47143214 | 0.12 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr8_+_85897951 | 0.12 |
ENSMUST00000076896.6
|
Cks1brt
|
CDC28 protein kinase 1b, retrogene |
| chr7_+_127503251 | 0.12 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr2_-_36782948 | 0.11 |
ENSMUST00000217215.3
|
Olfr353
|
olfactory receptor 353 |
| chr16_-_57113207 | 0.11 |
ENSMUST00000023434.15
ENSMUST00000120112.2 ENSMUST00000119407.8 |
Tmem30c
|
transmembrane protein 30C |
| chr15_+_99870714 | 0.11 |
ENSMUST00000230956.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chrX_+_158491589 | 0.10 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr9_+_72892786 | 0.10 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr5_+_124577952 | 0.10 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr3_-_27764571 | 0.09 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr2_+_71884943 | 0.09 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_102566717 | 0.09 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr3_+_105821450 | 0.09 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr10_+_74802996 | 0.09 |
ENSMUST00000037813.5
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr4_+_43441939 | 0.09 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1 |
| chr9_-_49710190 | 0.08 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_+_72892693 | 0.08 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr15_+_84553801 | 0.08 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr9_-_88601866 | 0.08 |
ENSMUST00000113110.5
|
Mthfsl
|
5, 10-methenyltetrahydrofolate synthetase-like |
| chr12_-_98700886 | 0.08 |
ENSMUST00000085116.4
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr10_+_127637015 | 0.07 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr19_-_4384029 | 0.07 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr2_-_79959178 | 0.07 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr9_+_89093210 | 0.07 |
ENSMUST00000118870.8
ENSMUST00000085256.8 |
Mthfs
|
5, 10-methenyltetrahydrofolate synthetase |
| chr3_-_95902949 | 0.07 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr1_+_90926443 | 0.06 |
ENSMUST00000189505.7
ENSMUST00000185531.7 ENSMUST00000068116.13 |
Lrrfip1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr5_-_66776095 | 0.06 |
ENSMUST00000162366.8
ENSMUST00000162994.8 ENSMUST00000159512.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr10_+_59715378 | 0.06 |
ENSMUST00000147914.8
ENSMUST00000146590.8 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr11_-_115494692 | 0.06 |
ENSMUST00000125097.2
ENSMUST00000019135.14 ENSMUST00000106508.10 |
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr19_+_26725589 | 0.06 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_68414401 | 0.06 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr18_+_38809771 | 0.06 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr5_-_124325213 | 0.06 |
ENSMUST00000161938.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr5_+_124621521 | 0.05 |
ENSMUST00000111453.2
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr10_-_95337783 | 0.05 |
ENSMUST00000075829.3
ENSMUST00000217777.2 ENSMUST00000218893.2 |
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr15_-_75963446 | 0.05 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr5_-_142594549 | 0.05 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr11_+_31822211 | 0.05 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr3_-_126792056 | 0.05 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr6_-_122317156 | 0.05 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr16_+_43056218 | 0.05 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_+_55443893 | 0.05 |
ENSMUST00000032627.5
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chrX_+_165127688 | 0.05 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr10_+_59715439 | 0.05 |
ENSMUST00000142819.8
ENSMUST00000020309.7 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr17_+_38231439 | 0.04 |
ENSMUST00000216440.2
|
Olfr128
|
olfactory receptor 128 |
| chr5_-_66775979 | 0.04 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr10_+_127595639 | 0.04 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr2_+_89842475 | 0.04 |
ENSMUST00000214382.2
ENSMUST00000217065.3 |
Olfr1263
|
olfactory receptor 1263 |
| chr14_-_20714634 | 0.04 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr19_+_46044972 | 0.04 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr7_-_23206631 | 0.04 |
ENSMUST00000227713.2
|
Vmn1r167
|
vomeronasal 1 receptor 167 |
| chr13_+_81034214 | 0.04 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr17_-_28841006 | 0.04 |
ENSMUST00000233353.2
|
Srpk1
|
serine/arginine-rich protein specific kinase 1 |
| chr5_-_3691453 | 0.03 |
ENSMUST00000140871.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr11_+_11414256 | 0.03 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr11_+_68393845 | 0.03 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr6_+_91881193 | 0.03 |
ENSMUST00000205686.2
|
4930590J08Rik
|
RIKEN cDNA 4930590J08 gene |
| chr7_+_12661337 | 0.03 |
ENSMUST00000045870.5
|
Rnf225
|
ring finger protein 225 |
| chr14_-_79718890 | 0.03 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr11_-_97886997 | 0.03 |
ENSMUST00000042971.16
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr1_-_139487951 | 0.03 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr8_+_104897074 | 0.03 |
ENSMUST00000164076.3
ENSMUST00000171018.8 ENSMUST00000167633.8 ENSMUST00000093245.13 ENSMUST00000212979.2 |
Bean1
|
brain expressed, associated with Nedd4, 1 |
| chr15_+_30172716 | 0.03 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr18_+_74349189 | 0.03 |
ENSMUST00000025444.8
|
Cxxc1
|
CXXC finger 1 (PHD domain) |
| chr14_-_10185481 | 0.03 |
ENSMUST00000225640.2
ENSMUST00000225294.2 ENSMUST00000224389.2 ENSMUST00000223927.2 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr6_+_97906760 | 0.03 |
ENSMUST00000101123.10
|
Mitf
|
melanogenesis associated transcription factor |
| chr3_+_3573084 | 0.03 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr9_+_66257747 | 0.03 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr13_-_67523832 | 0.03 |
ENSMUST00000225787.2
ENSMUST00000172266.8 ENSMUST00000057070.9 |
Zfp456
|
zinc finger protein 456 |
| chr9_-_21829385 | 0.03 |
ENSMUST00000128442.2
ENSMUST00000119055.8 ENSMUST00000122211.8 ENSMUST00000115351.10 |
Rab3d
|
RAB3D, member RAS oncogene family |
| chr10_-_14593935 | 0.03 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr11_+_53324126 | 0.02 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr13_+_108996606 | 0.02 |
ENSMUST00000177907.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr2_+_89708781 | 0.02 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr15_+_99870661 | 0.02 |
ENSMUST00000100206.4
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr4_-_116024788 | 0.02 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr2_-_79287095 | 0.02 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr10_+_80097290 | 0.02 |
ENSMUST00000156935.8
|
Dazap1
|
DAZ associated protein 1 |
| chr12_-_114710326 | 0.02 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr11_+_100211363 | 0.02 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr17_-_36280616 | 0.02 |
ENSMUST00000043757.15
|
Abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr8_-_70202576 | 0.02 |
ENSMUST00000130458.8
ENSMUST00000154063.2 |
Zfp963
|
zinc finger protein 963 |
| chr11_-_97673203 | 0.02 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr16_-_31133622 | 0.02 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr12_-_114878652 | 0.02 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr18_-_40352372 | 0.02 |
ENSMUST00000025364.6
|
Yipf5
|
Yip1 domain family, member 5 |
| chr5_-_135991117 | 0.02 |
ENSMUST00000111150.2
ENSMUST00000054895.4 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
| chr17_-_46798566 | 0.02 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chr12_-_114646685 | 0.02 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr4_+_85972125 | 0.02 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr9_+_106158212 | 0.02 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chr15_+_95698574 | 0.02 |
ENSMUST00000226793.2
|
Ano6
|
anoctamin 6 |
| chr7_+_30399208 | 0.02 |
ENSMUST00000013227.8
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr14_-_10185787 | 0.02 |
ENSMUST00000225871.2
|
Gm49355
|
predicted gene, 49355 |
| chr17_+_35345292 | 0.02 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr6_-_122317484 | 0.02 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr17_-_80885197 | 0.02 |
ENSMUST00000234602.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr8_+_72795062 | 0.01 |
ENSMUST00000215324.2
|
Olfr372
|
olfactory receptor 372 |
| chr11_+_95734028 | 0.01 |
ENSMUST00000107709.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr7_+_4928784 | 0.01 |
ENSMUST00000057612.9
|
Ssc5d
|
scavenger receptor cysteine rich family, 5 domains |
| chr3_-_10366229 | 0.01 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr14_-_10185523 | 0.01 |
ENSMUST00000223702.2
ENSMUST00000223762.2 ENSMUST00000112669.10 ENSMUST00000163392.3 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr17_-_80884839 | 0.01 |
ENSMUST00000234349.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr7_-_126303947 | 0.01 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr6_-_122317457 | 0.01 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr6_+_90439596 | 0.01 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr8_+_84293300 | 0.01 |
ENSMUST00000036996.6
|
Ndufb7
|
NADH:ubiquinone oxidoreductase subunit B7 |
| chr6_-_12749192 | 0.01 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr3_+_137770813 | 0.01 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr3_+_36530081 | 0.01 |
ENSMUST00000029268.7
|
1810062G17Rik
|
RIKEN cDNA 1810062G17 gene |
| chr11_-_98220466 | 0.01 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr11_-_75686874 | 0.01 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr17_+_38158374 | 0.01 |
ENSMUST00000213844.2
ENSMUST00000217487.2 |
Olfr126
|
olfactory receptor 126 |
| chr15_-_76010736 | 0.01 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr2_-_86970644 | 0.01 |
ENSMUST00000152758.4
|
Olfr1110
|
olfactory receptor 1110 |
| chr11_-_53313950 | 0.01 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr3_-_27764522 | 0.01 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr11_+_73051228 | 0.01 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_-_26187883 | 0.01 |
ENSMUST00000108308.10
ENSMUST00000075054.10 |
Nlgn1
|
neuroligin 1 |
| chr10_+_127685785 | 0.01 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr12_-_114815280 | 0.01 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr2_-_121637505 | 0.01 |
ENSMUST00000138157.8
|
Frmd5
|
FERM domain containing 5 |
| chr3_-_84167119 | 0.01 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chrY_-_3410148 | 0.01 |
ENSMUST00000190283.7
ENSMUST00000188091.7 ENSMUST00000169382.3 |
Gm21708
Gm21704
|
predicted gene, 21708 predicted gene, 21704 |
| chr9_+_37313193 | 0.01 |
ENSMUST00000214185.3
|
Robo4
|
roundabout guidance receptor 4 |
| chr17_+_38032699 | 0.01 |
ENSMUST00000207771.3
|
Olfr120
|
olfactory receptor 120 |
| chr6_+_90439544 | 0.01 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr4_-_118774662 | 0.01 |
ENSMUST00000071979.2
|
Olfr1329
|
olfactory receptor 1329 |
| chrY_+_2830680 | 0.01 |
ENSMUST00000171534.8
ENSMUST00000100360.5 ENSMUST00000179404.8 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chr5_+_105563605 | 0.01 |
ENSMUST00000112707.3
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr6_-_28421678 | 0.01 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr6_-_52203146 | 0.01 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr3_+_95341698 | 0.01 |
ENSMUST00000102749.11
ENSMUST00000090804.12 ENSMUST00000107161.8 ENSMUST00000107160.8 ENSMUST00000015666.17 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr4_-_118792037 | 0.01 |
ENSMUST00000081960.5
|
Olfr1328
|
olfactory receptor 1328 |
| chr10_-_128236317 | 0.01 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr11_+_58668915 | 0.01 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr7_+_17799863 | 0.01 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr3_-_46402355 | 0.01 |
ENSMUST00000195537.2
ENSMUST00000166505.7 ENSMUST00000195436.2 |
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr18_-_38417390 | 0.01 |
ENSMUST00000025311.8
|
Pcdh12
|
protocadherin 12 |
| chrY_+_2862139 | 0.01 |
ENSMUST00000189964.7
ENSMUST00000188114.2 |
Gm10256
|
predicted gene 10256 |
| chr17_-_33687660 | 0.01 |
ENSMUST00000234517.3
|
Gm4125
|
predicted gene 4125 |
| chr14_+_47535717 | 0.01 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr14_-_66191177 | 0.00 |
ENSMUST00000042046.5
|
Scara3
|
scavenger receptor class A, member 3 |
| chr6_-_116050081 | 0.00 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr8_+_11778039 | 0.00 |
ENSMUST00000110909.9
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr2_+_125876566 | 0.00 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr7_-_133203838 | 0.00 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr18_-_38417444 | 0.00 |
ENSMUST00000194012.2
|
Pcdh12
|
protocadherin 12 |
| chr15_-_103929556 | 0.00 |
ENSMUST00000226584.2
ENSMUST00000037685.9 |
Mucl2
|
mucin-like 2 |
| chr13_-_99584091 | 0.00 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chrX_+_75436956 | 0.00 |
ENSMUST00000101419.2
ENSMUST00000178974.2 |
Cldn34b4
|
claudin 34B4 |
| chr1_+_98348817 | 0.00 |
ENSMUST00000027575.13
ENSMUST00000160796.2 |
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr2_+_36827133 | 0.00 |
ENSMUST00000095021.2
|
Olfr356
|
olfactory receptor 356 |
| chr4_-_118314647 | 0.00 |
ENSMUST00000106375.2
ENSMUST00000168404.9 ENSMUST00000006556.11 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr3_+_28038283 | 0.00 |
ENSMUST00000067757.11
ENSMUST00000123539.8 |
Pld1
|
phospholipase D1 |
| chr12_-_91712783 | 0.00 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr10_+_127702326 | 0.00 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr16_-_44153288 | 0.00 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr9_+_37313287 | 0.00 |
ENSMUST00000115048.10
ENSMUST00000115046.9 ENSMUST00000102895.7 ENSMUST00000239486.2 |
Robo4
|
roundabout guidance receptor 4 |
| chr4_-_49681954 | 0.00 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr16_-_44153498 | 0.00 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr2_+_154042291 | 0.00 |
ENSMUST00000028987.7
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr14_+_63235512 | 0.00 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr7_+_142662931 | 0.00 |
ENSMUST00000187213.2
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1 |
| chr14_-_75830550 | 0.00 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr7_-_103420801 | 0.00 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chrY_-_2796205 | 0.00 |
ENSMUST00000187482.2
|
Gm4064
|
predicted gene 4064 |
| chrY_-_3306449 | 0.00 |
ENSMUST00000189592.7
|
Gm21677
|
predicted gene, 21677 |
| chrY_-_3378783 | 0.00 |
ENSMUST00000187277.7
|
Gm21704
|
predicted gene, 21704 |
| chr4_-_118314707 | 0.00 |
ENSMUST00000102671.10
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr7_-_42097503 | 0.00 |
ENSMUST00000032648.5
|
4933421I07Rik
|
RIKEN cDNA 4933421I07 gene |
| chr3_-_123029782 | 0.00 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr15_+_101013704 | 0.00 |
ENSMUST00000229954.2
|
Ankrd33
|
ankyrin repeat domain 33 |
| chrY_+_2900989 | 0.00 |
ENSMUST00000187842.7
|
Gm10352
|
predicted gene 10352 |
| chr9_+_86897590 | 0.00 |
ENSMUST00000058846.11
|
Ripply2
|
ripply transcriptional repressor 2 |
| chrY_-_3345329 | 0.00 |
ENSMUST00000186047.7
|
Gm21693
|
predicted gene, 21693 |
| chr2_+_86655007 | 0.00 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod2_Bhlha15_Bhlhe22_Olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046832 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:2000405 | negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |