Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
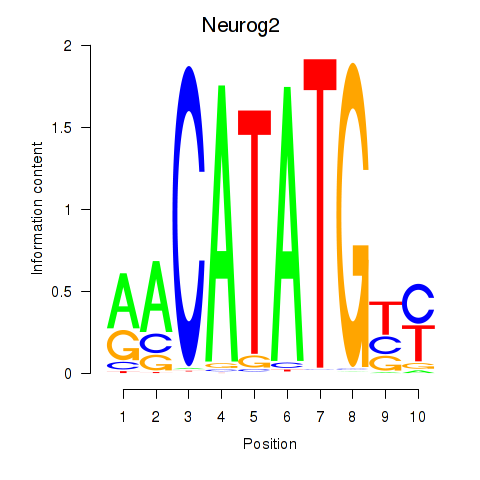
Results for Neurog2
Z-value: 1.64
Transcription factors associated with Neurog2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurog2
|
ENSMUSG00000027967.9 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurog2 | mm39_v1_chr3_+_127426783_127426789 | -0.99 | 1.0e-03 | Click! |
Activity profile of Neurog2 motif
Sorted Z-values of Neurog2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_60907698 | 1.36 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr7_-_126303887 | 1.33 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_+_49138278 | 1.13 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr7_-_126303689 | 1.09 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr18_+_60907668 | 1.09 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr10_+_55982969 | 0.98 |
ENSMUST00000063138.8
|
Msl3l2
|
MSL3 like 2 |
| chr17_-_26011357 | 0.97 |
ENSMUST00000236683.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr14_+_43951187 | 0.88 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr15_+_81912312 | 0.85 |
ENSMUST00000230729.2
|
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr3_-_105940130 | 0.79 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr11_-_84807164 | 0.75 |
ENSMUST00000103195.5
|
Znhit3
|
zinc finger, HIT type 3 |
| chr9_+_56344700 | 0.70 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr2_-_101479846 | 0.68 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr9_+_108437485 | 0.67 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr11_+_120123727 | 0.67 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr18_-_43610829 | 0.65 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_-_72441054 | 0.64 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr14_-_70945434 | 0.63 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chrX_+_35592006 | 0.62 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr7_-_28649094 | 0.62 |
ENSMUST00000148196.3
|
Actn4
|
actinin alpha 4 |
| chr9_+_66853343 | 0.61 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chrX_+_55500170 | 0.59 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr11_-_101308441 | 0.57 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr17_+_29899420 | 0.53 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chrX_+_36059274 | 0.50 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr6_+_17743581 | 0.50 |
ENSMUST00000000674.13
ENSMUST00000077080.9 |
St7
|
suppression of tumorigenicity 7 |
| chr10_+_128067964 | 0.50 |
ENSMUST00000125289.8
ENSMUST00000105242.8 |
Timeless
|
timeless circadian clock 1 |
| chr8_+_86219191 | 0.49 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr11_-_69811717 | 0.49 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr4_-_82768958 | 0.48 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr6_-_148732946 | 0.47 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr11_+_117123107 | 0.47 |
ENSMUST00000106354.9
|
Septin9
|
septin 9 |
| chr9_-_26910833 | 0.47 |
ENSMUST00000060513.8
ENSMUST00000120367.8 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chr8_+_123202935 | 0.47 |
ENSMUST00000146634.8
ENSMUST00000134127.2 |
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr2_+_163535925 | 0.45 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr11_-_103829040 | 0.45 |
ENSMUST00000133774.4
ENSMUST00000149642.3 |
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr2_-_71198091 | 0.42 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr7_-_64041996 | 0.42 |
ENSMUST00000032735.8
|
Mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr11_-_69811347 | 0.41 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr15_+_65682066 | 0.41 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr10_+_128067934 | 0.40 |
ENSMUST00000055539.11
ENSMUST00000105244.8 ENSMUST00000105243.9 |
Timeless
|
timeless circadian clock 1 |
| chrX_+_7445806 | 0.40 |
ENSMUST00000234363.2
ENSMUST00000235116.2 ENSMUST00000115739.9 ENSMUST00000234574.2 ENSMUST00000115740.9 |
Foxp3
|
forkhead box P3 |
| chr15_-_85695855 | 0.40 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr6_+_51521493 | 0.40 |
ENSMUST00000179365.8
ENSMUST00000114439.8 |
Snx10
|
sorting nexin 10 |
| chr17_-_26011241 | 0.39 |
ENSMUST00000237093.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr13_+_21364330 | 0.39 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr5_+_3393893 | 0.39 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr2_+_173918715 | 0.38 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr6_+_124489364 | 0.38 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr4_-_12087911 | 0.38 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67 |
| chr1_+_177272297 | 0.38 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_+_108162985 | 0.37 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr9_+_111011388 | 0.37 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr4_-_115911053 | 0.37 |
ENSMUST00000030475.3
|
Nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr15_+_51741138 | 0.37 |
ENSMUST00000136129.2
|
Utp23
|
UTP23 small subunit processome component |
| chr10_+_110756031 | 0.36 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr9_-_119151428 | 0.36 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr7_+_127503812 | 0.36 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr1_+_135945798 | 0.36 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr1_+_135945705 | 0.36 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr1_+_177272215 | 0.34 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_+_84081411 | 0.34 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr9_+_108539296 | 0.33 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr19_+_37184927 | 0.33 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr18_-_36587573 | 0.33 |
ENSMUST00000025204.7
ENSMUST00000237792.2 |
Pfdn1
|
prefoldin 1 |
| chr11_+_32592707 | 0.33 |
ENSMUST00000109366.8
ENSMUST00000093205.13 ENSMUST00000076383.8 |
Fbxw11
|
F-box and WD-40 domain protein 11 |
| chr19_+_45991907 | 0.32 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr7_+_30014235 | 0.32 |
ENSMUST00000054594.15
ENSMUST00000177078.8 ENSMUST00000176504.8 ENSMUST00000176304.8 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr2_+_164298854 | 0.31 |
ENSMUST00000109352.8
|
Sys1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr2_+_127429125 | 0.31 |
ENSMUST00000028852.13
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr8_+_123202882 | 0.30 |
ENSMUST00000116412.8
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr8_+_95564949 | 0.29 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr6_-_52203146 | 0.29 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr4_+_11579648 | 0.28 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr2_-_25391729 | 0.28 |
ENSMUST00000015227.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr6_-_83010402 | 0.28 |
ENSMUST00000089651.6
|
Dok1
|
docking protein 1 |
| chr5_+_90638580 | 0.28 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr14_+_74969737 | 0.27 |
ENSMUST00000022573.17
ENSMUST00000175712.8 ENSMUST00000176957.8 |
Esd
|
esterase D/formylglutathione hydrolase |
| chr8_-_34614187 | 0.27 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr2_+_157579321 | 0.27 |
ENSMUST00000029178.7
|
Ctnnbl1
|
catenin, beta like 1 |
| chr14_-_10185523 | 0.27 |
ENSMUST00000223702.2
ENSMUST00000223762.2 ENSMUST00000112669.10 ENSMUST00000163392.3 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr7_+_127187910 | 0.27 |
ENSMUST00000205694.2
ENSMUST00000033088.8 ENSMUST00000206914.2 |
Rnf40
|
ring finger protein 40 |
| chr12_-_79027531 | 0.26 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr16_+_16031182 | 0.24 |
ENSMUST00000039408.3
|
Pkp2
|
plakophilin 2 |
| chr19_-_53577499 | 0.24 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr8_+_95393228 | 0.23 |
ENSMUST00000034228.16
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr9_-_71803354 | 0.22 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr2_+_75662511 | 0.22 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chr1_-_131441962 | 0.21 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr6_-_148732893 | 0.20 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr7_+_28488380 | 0.20 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr7_-_44397730 | 0.20 |
ENSMUST00000118162.8
ENSMUST00000140599.9 ENSMUST00000120798.8 |
Zfp473
|
zinc finger protein 473 |
| chr2_+_69500444 | 0.20 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr15_-_85695810 | 0.20 |
ENSMUST00000144067.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr10_-_116385007 | 0.19 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr19_+_20579322 | 0.19 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr14_-_70405288 | 0.19 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr4_-_108158242 | 0.19 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr2_+_57887896 | 0.18 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr15_-_91075933 | 0.18 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr15_-_77854711 | 0.18 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr1_-_150268470 | 0.17 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr13_+_16189041 | 0.17 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr17_+_29042544 | 0.16 |
ENSMUST00000140587.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr9_+_111011327 | 0.16 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_59442378 | 0.16 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_+_88128323 | 0.16 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr14_-_10185481 | 0.16 |
ENSMUST00000225640.2
ENSMUST00000225294.2 ENSMUST00000224389.2 ENSMUST00000223927.2 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr11_-_101316156 | 0.15 |
ENSMUST00000103102.10
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr4_+_8535604 | 0.15 |
ENSMUST00000060232.8
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr6_-_54941673 | 0.14 |
ENSMUST00000203837.2
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr2_-_174188505 | 0.14 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr9_-_21829385 | 0.14 |
ENSMUST00000128442.2
ENSMUST00000119055.8 ENSMUST00000122211.8 ENSMUST00000115351.10 |
Rab3d
|
RAB3D, member RAS oncogene family |
| chr17_-_34822649 | 0.13 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr2_-_29142965 | 0.13 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr7_+_130633776 | 0.13 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr5_-_90487583 | 0.13 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr17_+_38104420 | 0.12 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr6_-_48685108 | 0.12 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chr9_-_60557076 | 0.12 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr4_+_9269285 | 0.12 |
ENSMUST00000038841.14
|
Clvs1
|
clavesin 1 |
| chr11_+_95734419 | 0.12 |
ENSMUST00000107708.2
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr1_+_6800275 | 0.12 |
ENSMUST00000043578.13
ENSMUST00000139756.8 ENSMUST00000131467.8 ENSMUST00000150761.8 ENSMUST00000151281.8 |
St18
|
suppression of tumorigenicity 18 |
| chr2_+_109522781 | 0.11 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr11_+_117157024 | 0.11 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chrX_-_7054952 | 0.11 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr7_+_44397837 | 0.11 |
ENSMUST00000002275.15
|
Vrk3
|
vaccinia related kinase 3 |
| chrX_+_106192510 | 0.11 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr1_-_135040044 | 0.11 |
ENSMUST00000086432.7
|
Ptprv
|
protein tyrosine phosphatase, receptor type, V |
| chr17_+_3532455 | 0.10 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr5_-_143314938 | 0.10 |
ENSMUST00000162941.8
ENSMUST00000159941.8 ENSMUST00000162066.8 ENSMUST00000162358.8 ENSMUST00000001900.9 |
Zdhhc4
|
zinc finger, DHHC domain containing 4 |
| chr10_-_127358300 | 0.10 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_-_37153436 | 0.10 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr15_-_77854988 | 0.10 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr11_-_69696428 | 0.09 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr17_+_47083561 | 0.09 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr10_-_5019044 | 0.09 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_164763091 | 0.09 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr10_-_95509251 | 0.09 |
ENSMUST00000218771.2
ENSMUST00000099328.2 |
Anapc15-ps
|
anaphase promoting complex C subunit 15, pseudogene |
| chrX_+_140258381 | 0.09 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr9_+_26910995 | 0.09 |
ENSMUST00000213770.2
ENSMUST00000213683.2 ENSMUST00000039161.10 |
Thyn1
|
thymocyte nuclear protein 1 |
| chr2_-_131953359 | 0.09 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr7_-_44397473 | 0.08 |
ENSMUST00000120074.8
|
Zfp473
|
zinc finger protein 473 |
| chr17_+_38104635 | 0.08 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr2_+_85597442 | 0.08 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr2_+_155224105 | 0.08 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr10_-_127358231 | 0.08 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_126303947 | 0.08 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr1_+_180762587 | 0.07 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr12_-_115276219 | 0.07 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr11_+_78079562 | 0.07 |
ENSMUST00000108322.9
|
Rab34
|
RAB34, member RAS oncogene family |
| chr19_+_5074070 | 0.07 |
ENSMUST00000025826.7
ENSMUST00000237371.2 ENSMUST00000235416.2 |
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr19_-_33764859 | 0.07 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr13_-_101829132 | 0.07 |
ENSMUST00000035532.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr17_-_37404764 | 0.07 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr9_+_19620729 | 0.06 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr9_-_110571645 | 0.06 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr12_-_114073050 | 0.06 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr4_-_110149916 | 0.06 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr17_+_18108102 | 0.06 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr12_-_114901026 | 0.06 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr6_-_30680502 | 0.06 |
ENSMUST00000133373.8
|
Cep41
|
centrosomal protein 41 |
| chr10_+_97442727 | 0.05 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr2_-_86276348 | 0.05 |
ENSMUST00000216165.2
|
Olfr1065
|
olfactory receptor 1065 |
| chr17_-_27842412 | 0.05 |
ENSMUST00000025050.13
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr9_+_58489523 | 0.05 |
ENSMUST00000177292.8
ENSMUST00000085651.12 ENSMUST00000176557.8 ENSMUST00000114121.11 ENSMUST00000177064.8 |
Nptn
|
neuroplastin |
| chr11_-_68743944 | 0.05 |
ENSMUST00000018880.14
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr2_-_110983281 | 0.05 |
ENSMUST00000132464.2
|
Gm15130
|
predicted gene 15130 |
| chrX_+_163156359 | 0.05 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr3_-_59127571 | 0.05 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr2_+_87576198 | 0.05 |
ENSMUST00000217572.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr13_+_67980849 | 0.05 |
ENSMUST00000221620.2
|
Gm10037
|
predicted gene 10037 |
| chr3_-_123029745 | 0.05 |
ENSMUST00000106426.8
|
Synpo2
|
synaptopodin 2 |
| chr9_+_19691579 | 0.05 |
ENSMUST00000212363.2
ENSMUST00000215112.3 |
Olfr58
|
olfactory receptor 58 |
| chr9_+_39580355 | 0.05 |
ENSMUST00000073433.2
|
Olfr963
|
olfactory receptor 963 |
| chr8_+_95393349 | 0.05 |
ENSMUST00000109527.6
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr6_+_68657317 | 0.05 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr3_-_123029782 | 0.05 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr9_-_18730822 | 0.05 |
ENSMUST00000215380.3
|
Olfr828
|
olfactory receptor 828 |
| chr18_-_35781422 | 0.05 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr19_+_13610308 | 0.04 |
ENSMUST00000053113.5
|
Olfr1489
|
olfactory receptor 1489 |
| chr13_-_67547858 | 0.04 |
ENSMUST00000224684.2
ENSMUST00000181071.8 ENSMUST00000109732.3 ENSMUST00000224825.2 |
Zfp429
|
zinc finger protein 429 |
| chr17_-_27842237 | 0.04 |
ENSMUST00000062397.13
ENSMUST00000176876.8 ENSMUST00000146321.3 |
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr6_+_57186843 | 0.04 |
ENSMUST00000078885.2
|
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr2_+_163444214 | 0.04 |
ENSMUST00000171696.8
ENSMUST00000109408.10 |
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr6_+_41331039 | 0.04 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr2_+_85805557 | 0.04 |
ENSMUST00000082191.5
|
Olfr1029
|
olfactory receptor 1029 |
| chr2_+_111491764 | 0.04 |
ENSMUST00000213511.2
ENSMUST00000207228.3 ENSMUST00000208983.2 |
Olfr1299
Gm44840
|
olfactory receptor 1299 predicted gene 44840 |
| chrY_-_40270796 | 0.04 |
ENSMUST00000177713.2
|
Gm21865
|
predicted gene, 21865 |
| chr19_-_53360197 | 0.04 |
ENSMUST00000086887.2
|
Gm10197
|
predicted gene 10197 |
| chr3_+_90434160 | 0.04 |
ENSMUST00000199538.5
ENSMUST00000164481.7 ENSMUST00000167598.6 |
S100a14
|
S100 calcium binding protein A14 |
| chr6_-_139478919 | 0.04 |
ENSMUST00000170650.3
|
Rergl
|
RERG/RAS-like |
| chrX_-_52610946 | 0.04 |
ENSMUST00000123034.3
|
4933416I08Rik
|
RIKEN cDNA 4933416I08 gene |
| chr9_-_38971960 | 0.04 |
ENSMUST00000055567.3
|
Olfr937
|
olfactory receptor 937 |
| chr9_-_70328816 | 0.04 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr2_-_93787441 | 0.04 |
ENSMUST00000099689.5
|
Gm13889
|
predicted gene 13889 |
| chr13_+_67979225 | 0.03 |
ENSMUST00000221266.2
|
Gm10037
|
predicted gene 10037 |
| chr11_+_53324126 | 0.03 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr8_+_10299288 | 0.03 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr2_+_85838122 | 0.03 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurog2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.3 | 1.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 0.8 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.9 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.9 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.7 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.4 | GO:0002849 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.4 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.1 | 0.5 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.4 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.5 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 3.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:2000412 | immunoglobulin production in mucosal tissue(GO:0002426) positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.4 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.2 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 2.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.0 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 3.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 2.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.1 | 0.3 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.8 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 2.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |