|
chr9_+_3000922
Show fit
|
15.09 |
ENSMUST00000151376.3
|
Gm10722
|
predicted gene 10722
|
|
chr9_+_3023547
Show fit
|
14.82 |
ENSMUST00000099046.4
|
Gm10718
|
predicted gene 10718
|
|
chr9_+_3013140
Show fit
|
14.29 |
ENSMUST00000143083.3
|
Gm10721
|
predicted gene 10721
|
|
chr9_+_3027439
Show fit
|
13.58 |
ENSMUST00000179982.2
ENSMUST00000177875.8
|
Gm10717
|
predicted gene 10717
|
|
chr9_+_3017408
Show fit
|
12.16 |
ENSMUST00000099049.4
|
Gm10719
|
predicted gene 10719
|
|
chr9_+_3004457
Show fit
|
12.15 |
ENSMUST00000178348.2
|
Gm11168
|
predicted gene 11168
|
|
chr9_+_3025417
Show fit
|
12.01 |
ENSMUST00000075573.7
|
Gm10717
|
predicted gene 10717
|
|
chr9_+_3034599
Show fit
|
11.63 |
ENSMUST00000178641.2
|
Gm17535
|
predicted gene, 17535
|
|
chr9_+_3005126
Show fit
|
10.99 |
ENSMUST00000179881.2
|
Gm11168
|
predicted gene 11168
|
|
chr9_+_3018753
Show fit
|
9.19 |
ENSMUST00000179272.2
|
Gm10719
|
predicted gene 10719
|
|
chr13_-_23945189
Show fit
|
7.83 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1
|
|
chr9_+_3032001
Show fit
|
7.27 |
ENSMUST00000099056.4
ENSMUST00000179839.8
|
Gm10717
|
predicted gene 10717
|
|
chr9_+_3036877
Show fit
|
7.26 |
ENSMUST00000155807.3
|
Gm10715
|
predicted gene 10715
|
|
chr9_+_3037111
Show fit
|
7.08 |
ENSMUST00000177969.2
|
Gm10715
|
predicted gene 10715
|
|
chr9_+_3015654
Show fit
|
6.75 |
ENSMUST00000099050.4
|
Gm10720
|
predicted gene 10720
|
|
chr2_-_98497609
Show fit
|
5.52 |
ENSMUST00000099683.2
|
Gm10800
|
predicted gene 10800
|
|
chr12_+_33364288
Show fit
|
3.56 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr7_-_85895409
Show fit
|
3.53 |
ENSMUST00000165771.2
ENSMUST00000233075.2
ENSMUST00000233317.2
ENSMUST00000233312.2
ENSMUST00000233928.2
ENSMUST00000232799.2
ENSMUST00000233744.2
|
Vmn2r76
|
vomeronasal 2, receptor 76
|
|
chr2_+_98492576
Show fit
|
3.50 |
ENSMUST00000099684.4
|
Gm10801
|
predicted gene 10801
|
|
chr15_+_6609322
Show fit
|
3.13 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein
|
|
chr6_-_97408367
Show fit
|
3.10 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B
|
|
chr1_+_107456731
Show fit
|
3.06 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr7_-_126224848
Show fit
|
2.84 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1
|
|
chr10_+_40505985
Show fit
|
2.81 |
ENSMUST00000019977.8
ENSMUST00000214102.2
ENSMUST00000213503.2
ENSMUST00000213442.2
ENSMUST00000216830.2
|
Ddo
|
D-aspartate oxidase
|
|
chr13_+_23922783
Show fit
|
2.80 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member
|
|
chr11_-_11577824
Show fit
|
2.52 |
ENSMUST00000081896.5
|
4930512M02Rik
|
RIKEN cDNA 4930512M02 gene
|
|
chrX_-_9335525
Show fit
|
2.48 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr2_+_30127692
Show fit
|
2.41 |
ENSMUST00000113654.8
ENSMUST00000095078.3
|
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A
|
|
chrX_+_158410229
Show fit
|
2.37 |
ENSMUST00000112456.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_34044940
Show fit
|
2.30 |
ENSMUST00000187486.7
ENSMUST00000182697.8
|
Dst
|
dystonin
|
|
chr2_-_87868043
Show fit
|
2.16 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73
|
|
chr7_+_100435548
Show fit
|
2.02 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A
|
|
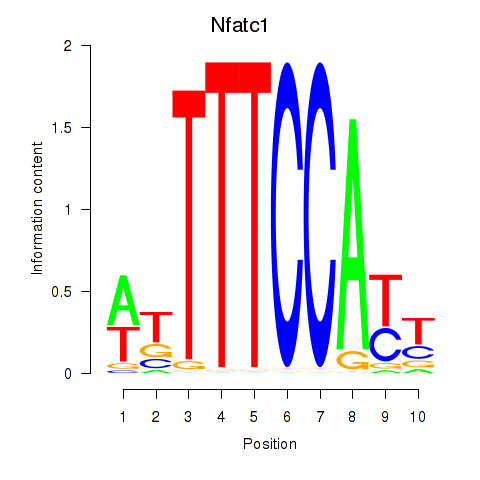
chr18_-_80756273
Show fit
|
1.98 |
ENSMUST00000078049.12
ENSMUST00000236711.2
|
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1
|
|
chr11_-_79421397
Show fit
|
1.96 |
ENSMUST00000103236.4
ENSMUST00000170799.8
ENSMUST00000170422.4
|
Evi2a
Evi2
|
ecotropic viral integration site 2a
ecotropic viral integration site 2
|
|
chr9_-_29874401
Show fit
|
1.95 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin
|
|
chrX_+_158410528
Show fit
|
1.94 |
ENSMUST00000073094.10
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr10_-_12689345
Show fit
|
1.88 |
ENSMUST00000217899.2
|
Utrn
|
utrophin
|
|
chr11_-_73881233
Show fit
|
1.88 |
ENSMUST00000214334.2
ENSMUST00000117510.4
|
Olfr398
|
olfactory receptor 398
|
|
chr2_-_87838612
Show fit
|
1.87 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160
|
|
chr6_-_57991216
Show fit
|
1.80 |
ENSMUST00000228951.2
|
Vmn1r26
|
vomeronasal 1 receptor 26
|
|
chr7_+_101619897
Show fit
|
1.79 |
ENSMUST00000211272.2
|
Numa1
|
nuclear mitotic apparatus protein 1
|
|
chr6_-_28831746
Show fit
|
1.76 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4
|
|
chr14_-_50586329
Show fit
|
1.74 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735
|
|
chr13_+_41013230
Show fit
|
1.73 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme
|
|
chr4_-_135080471
Show fit
|
1.69 |
ENSMUST00000084846.12
ENSMUST00000136342.9
ENSMUST00000105861.8
|
Srrm1
|
serine/arginine repetitive matrix 1
|
|
chr19_+_10502612
Show fit
|
1.69 |
ENSMUST00000237321.2
ENSMUST00000038379.5
|
Cpsf7
|
cleavage and polyadenylation specific factor 7
|
|
chr11_-_70545450
Show fit
|
1.58 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1
|
|
chr7_+_102954855
Show fit
|
1.56 |
ENSMUST00000214577.2
|
Olfr596
|
olfactory receptor 596
|
|
chr7_-_11414074
Show fit
|
1.56 |
ENSMUST00000227010.2
ENSMUST00000209638.3
|
Vmn1r72
|
vomeronasal 1 receptor 72
|
|
chr15_-_58828321
Show fit
|
1.55 |
ENSMUST00000228067.2
|
Mtss1
|
MTSS I-BAR domain containing 1
|
|
chr16_+_65317389
Show fit
|
1.52 |
ENSMUST00000176330.8
ENSMUST00000004964.15
ENSMUST00000176038.8
|
Pou1f1
|
POU domain, class 1, transcription factor 1
|
|
chr19_+_13339600
Show fit
|
1.51 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467
|
|
chr11_+_29413734
Show fit
|
1.51 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A
|
|
chr6_+_149211225
Show fit
|
1.49 |
ENSMUST00000185930.2
|
Resf1
|
retroelement silencing factor 1
|
|
chrX_-_37723943
Show fit
|
1.49 |
ENSMUST00000058265.8
|
C1galt1c1
|
C1GALT1-specific chaperone 1
|
|
chr1_-_60082246
Show fit
|
1.48 |
ENSMUST00000027172.13
ENSMUST00000191251.7
|
Ica1l
|
islet cell autoantigen 1-like
|
|
chr2_-_111779785
Show fit
|
1.41 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307
|
|
chr19_+_10502679
Show fit
|
1.37 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7
|
|
chr17_-_35340189
Show fit
|
1.37 |
ENSMUST00000025246.13
ENSMUST00000173114.8
|
Csnk2b
|
casein kinase 2, beta polypeptide
|
|
chr12_-_99359265
Show fit
|
1.35 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3
|
|
chr18_+_36693646
Show fit
|
1.34 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1
|
|
chr18_+_36693024
Show fit
|
1.33 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1
|
|
chr5_-_90031180
Show fit
|
1.32 |
ENSMUST00000198151.2
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3
|
|
chr6_+_15185202
Show fit
|
1.32 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2
|
|
chr5_+_138228074
Show fit
|
1.29 |
ENSMUST00000159798.8
ENSMUST00000159964.2
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5
|
|
chr2_+_89804937
Show fit
|
1.28 |
ENSMUST00000214630.2
ENSMUST00000111512.10
ENSMUST00000144710.3
ENSMUST00000216678.2
|
Olfr1260
|
olfactory receptor 1260
|
|
chr9_+_123901979
Show fit
|
1.28 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2
|
|
chr9_+_100525807
Show fit
|
1.27 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1
|
|
chr1_+_88022776
Show fit
|
1.26 |
ENSMUST00000150634.8
ENSMUST00000058237.14
|
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C
|
|
chr6_+_149210889
Show fit
|
1.23 |
ENSMUST00000130664.8
ENSMUST00000046689.13
|
Resf1
|
retroelement silencing factor 1
|
|
chr3_-_64752138
Show fit
|
1.19 |
ENSMUST00000177146.2
ENSMUST00000161972.10
|
Gm26939
Vmn2r7
|
predicted gene, 26939
vomeronasal 2, receptor 7
|
|
chr7_+_106737534
Show fit
|
1.18 |
ENSMUST00000213367.3
ENSMUST00000214819.3
ENSMUST00000216871.3
ENSMUST00000215284.3
ENSMUST00000209942.2
|
Olfr716
|
olfactory receptor 716
|
|
chr19_-_10502468
Show fit
|
1.18 |
ENSMUST00000025570.8
ENSMUST00000236455.2
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr9_-_123691077
Show fit
|
1.17 |
ENSMUST00000182350.3
|
Xcr1
|
chemokine (C motif) receptor 1
|
|
chr16_+_43067641
Show fit
|
1.16 |
ENSMUST00000079441.13
ENSMUST00000114691.8
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr15_-_9748863
Show fit
|
1.13 |
ENSMUST00000159368.2
ENSMUST00000159093.8
ENSMUST00000162780.8
ENSMUST00000160236.8
ENSMUST00000208854.2
ENSMUST00000041840.14
|
Spef2
|
sperm flagellar 2
|
|
chr14_+_35816874
Show fit
|
1.13 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene
|
|
chr18_+_31768126
Show fit
|
1.12 |
ENSMUST00000234846.2
ENSMUST00000234772.2
|
Sap130
|
Sin3A associated protein
|
|
chr6_+_149210941
Show fit
|
1.10 |
ENSMUST00000190785.7
ENSMUST00000189932.7
ENSMUST00000100765.11
|
Resf1
|
retroelement silencing factor 1
|
|
chr15_-_83135920
Show fit
|
1.10 |
ENSMUST00000229687.2
ENSMUST00000164614.3
ENSMUST00000049530.14
|
A4galt
|
alpha 1,4-galactosyltransferase
|
|
chr13_-_41513215
Show fit
|
1.09 |
ENSMUST00000224803.2
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr3_+_20043315
Show fit
|
1.08 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin
|
|
chr9_-_40366966
Show fit
|
1.07 |
ENSMUST00000165104.8
ENSMUST00000045682.7
|
Gramd1b
|
GRAM domain containing 1B
|
|
chrX_+_152281200
Show fit
|
1.06 |
ENSMUST00000060714.10
|
Ubqln2
|
ubiquilin 2
|
|
chr10_-_129524028
Show fit
|
1.06 |
ENSMUST00000203785.3
ENSMUST00000217576.2
|
Olfr802
|
olfactory receptor 802
|
|
chr14_+_15749522
Show fit
|
1.05 |
ENSMUST00000171987.2
|
Gm8050
|
predicted gene 8050
|
|
chr13_+_44884740
Show fit
|
1.05 |
ENSMUST00000173246.8
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr13_-_105191403
Show fit
|
1.04 |
ENSMUST00000063551.7
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr2_-_22930149
Show fit
|
1.04 |
ENSMUST00000091394.13
ENSMUST00000093171.13
|
Abi1
|
abl interactor 1
|
|
chr19_+_10366450
Show fit
|
1.02 |
ENSMUST00000073899.6
|
Syt7
|
synaptotagmin VII
|
|
chr7_+_75292971
Show fit
|
0.99 |
ENSMUST00000207998.2
|
Akap13
|
A kinase (PRKA) anchor protein 13
|
|
chr7_+_107497109
Show fit
|
0.98 |
ENSMUST00000209670.4
ENSMUST00000216937.3
|
Olfr472
|
olfactory receptor 472
|
|
chr2_+_88217406
Show fit
|
0.98 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178
|
|
chr7_+_30252687
Show fit
|
0.97 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr17_+_15262510
Show fit
|
0.94 |
ENSMUST00000226561.2
|
Ermard
|
ER membrane associated RNA degradation
|
|
chr14_-_20133246
Show fit
|
0.93 |
ENSMUST00000059666.6
|
Saysd1
|
SAYSVFN motif domain containing 1
|
|
chr4_-_148021159
Show fit
|
0.92 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr2_-_113588983
Show fit
|
0.92 |
ENSMUST00000099575.4
|
Grem1
|
gremlin 1, DAN family BMP antagonist
|
|
chr11_-_65636651
Show fit
|
0.89 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4
|
|
chr15_-_41733099
Show fit
|
0.88 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein
|
|
chr7_+_110372860
Show fit
|
0.88 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr2_-_73360069
Show fit
|
0.87 |
ENSMUST00000094681.11
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr1_-_84816379
Show fit
|
0.87 |
ENSMUST00000187818.2
|
Trip12
|
thyroid hormone receptor interactor 12
|
|
chr13_+_83672389
Show fit
|
0.85 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr10_-_22025169
Show fit
|
0.85 |
ENSMUST00000179054.8
ENSMUST00000069372.7
|
E030030I06Rik
|
RIKEN cDNA E030030I06 gene
|
|
chrY_-_2663658
Show fit
|
0.85 |
ENSMUST00000091178.2
|
Sry
|
sex determining region of Chr Y
|
|
chr7_-_29553079
Show fit
|
0.85 |
ENSMUST00000108223.8
|
Zfp940
|
zinc finger protein 940
|
|
chr14_-_20027279
Show fit
|
0.84 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr11_+_44508137
Show fit
|
0.82 |
ENSMUST00000109268.2
ENSMUST00000101326.10
ENSMUST00000081265.12
|
Ebf1
|
early B cell factor 1
|
|
chr7_+_63835285
Show fit
|
0.81 |
ENSMUST00000206263.2
ENSMUST00000206107.2
ENSMUST00000205731.2
ENSMUST00000206706.2
ENSMUST00000205690.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1
|
|
chr9_+_100525501
Show fit
|
0.81 |
ENSMUST00000146312.8
ENSMUST00000129269.8
|
Stag1
|
stromal antigen 1
|
|
chr11_+_49410475
Show fit
|
0.79 |
ENSMUST00000204706.3
|
Olfr1383
|
olfactory receptor 1383
|
|
chr7_-_140087224
Show fit
|
0.79 |
ENSMUST00000209873.2
ENSMUST00000064392.8
ENSMUST00000215768.2
ENSMUST00000215340.2
|
Olfr536
|
olfactory receptor 536
|
|
chr11_-_102907991
Show fit
|
0.79 |
ENSMUST00000021313.9
|
Dcakd
|
dephospho-CoA kinase domain containing
|
|
chr5_+_117979899
Show fit
|
0.78 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal
|
|
chr2_-_22930104
Show fit
|
0.78 |
ENSMUST00000153931.8
ENSMUST00000123948.8
|
Abi1
|
abl interactor 1
|
|
chr1_+_24717722
Show fit
|
0.78 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1
|
|
chr8_+_34582184
Show fit
|
0.76 |
ENSMUST00000095345.5
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4
|
|
chr5_+_104350475
Show fit
|
0.76 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1
|
|
chr15_-_66673425
Show fit
|
0.76 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor
|
|
chr8_+_66838927
Show fit
|
0.75 |
ENSMUST00000039540.12
ENSMUST00000110253.3
|
Marchf1
|
membrane associated ring-CH-type finger 1
|
|
chr2_-_77000936
Show fit
|
0.75 |
ENSMUST00000164114.9
ENSMUST00000049544.14
|
Ccdc141
|
coiled-coil domain containing 141
|
|
chr11_-_98329782
Show fit
|
0.73 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1
|
|
chr17_-_37627945
Show fit
|
0.72 |
ENSMUST00000217590.2
|
Olfr102
|
olfactory receptor 102
|
|
chr14_+_25842565
Show fit
|
0.71 |
ENSMUST00000022416.15
|
Anxa11
|
annexin A11
|
|
chr3_-_113423774
Show fit
|
0.71 |
ENSMUST00000092154.10
ENSMUST00000106536.8
ENSMUST00000106535.2
|
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3
|
|
chr3_-_88366351
Show fit
|
0.70 |
ENSMUST00000165898.8
ENSMUST00000127436.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chrX_-_16777913
Show fit
|
0.70 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human)
|
|
chr14_-_54924062
Show fit
|
0.69 |
ENSMUST00000147714.8
|
Acin1
|
apoptotic chromatin condensation inducer 1
|
|
chr13_+_83672654
Show fit
|
0.69 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chrX_-_125723491
Show fit
|
0.68 |
ENSMUST00000081074.5
|
4932411N23Rik
|
RIKEN cDNA 4932411N23 gene
|
|
chr4_+_86666764
Show fit
|
0.68 |
ENSMUST00000045512.15
ENSMUST00000082026.14
|
Dennd4c
|
DENN/MADD domain containing 4C
|
|
chr9_+_51959534
Show fit
|
0.67 |
ENSMUST00000061352.11
|
Rdx
|
radixin
|
|
chr15_+_101152078
Show fit
|
0.66 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr7_-_45159790
Show fit
|
0.66 |
ENSMUST00000211765.2
|
Nucb1
|
nucleobindin 1
|
|
chr3_+_90520408
Show fit
|
0.65 |
ENSMUST00000198128.2
|
S100a6
|
S100 calcium binding protein A6 (calcyclin)
|
|
chr8_-_105350898
Show fit
|
0.64 |
ENSMUST00000212882.2
ENSMUST00000163783.4
|
Cdh16
|
cadherin 16
|
|
chr3_-_67371161
Show fit
|
0.64 |
ENSMUST00000058981.3
|
Lxn
|
latexin
|
|
chr3_+_123061094
Show fit
|
0.63 |
ENSMUST00000047923.12
ENSMUST00000200333.2
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae)
|
|
chr15_-_99717956
Show fit
|
0.63 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1
|
|
chr18_+_69726431
Show fit
|
0.63 |
ENSMUST00000201091.4
ENSMUST00000201037.4
ENSMUST00000114977.5
|
Tcf4
|
transcription factor 4
|
|
chr1_-_155912216
Show fit
|
0.62 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1
|
|
chr13_-_93774469
Show fit
|
0.62 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase
|
|
chrX_-_104919201
Show fit
|
0.62 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler
|
|
chr9_+_100525637
Show fit
|
0.61 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1
|
|
chr13_+_83652150
Show fit
|
0.61 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr2_-_22930188
Show fit
|
0.61 |
ENSMUST00000114544.10
ENSMUST00000139038.8
ENSMUST00000126112.8
ENSMUST00000178908.2
ENSMUST00000078977.14
ENSMUST00000140164.8
ENSMUST00000149719.8
|
Abi1
|
abl interactor 1
|
|
chr18_+_64387428
Show fit
|
0.61 |
ENSMUST00000025477.15
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr2_-_88534814
Show fit
|
0.60 |
ENSMUST00000216928.2
ENSMUST00000216977.2
|
Olfr1196
|
olfactory receptor 1196
|
|
chr15_+_61857390
Show fit
|
0.60 |
ENSMUST00000159327.2
ENSMUST00000167731.8
|
Myc
|
myelocytomatosis oncogene
|
|
chr9_-_37627519
Show fit
|
0.59 |
ENSMUST00000215727.2
ENSMUST00000211952.3
|
Olfr160
|
olfactory receptor 160
|
|
chr6_-_41751648
Show fit
|
0.59 |
ENSMUST00000214752.2
|
Olfr459
|
olfactory receptor 459
|
|
chr9_+_123902143
Show fit
|
0.59 |
ENSMUST00000168841.3
ENSMUST00000055918.7
|
Ccr2
|
chemokine (C-C motif) receptor 2
|
|
chr14_+_52254341
Show fit
|
0.59 |
ENSMUST00000228408.2
ENSMUST00000227295.2
|
Tmem253
|
transmembrane protein 253
|
|
chr8_+_107237483
Show fit
|
0.58 |
ENSMUST00000080797.8
|
Cdh3
|
cadherin 3
|
|
chr4_+_155709275
Show fit
|
0.58 |
ENSMUST00000067081.10
ENSMUST00000105600.8
ENSMUST00000105598.2
|
Cdk11b
|
cyclin-dependent kinase 11B
|
|
chr13_-_67831586
Show fit
|
0.56 |
ENSMUST00000137496.10
ENSMUST00000175678.2
ENSMUST00000175821.8
ENSMUST00000125495.10
|
Zfp738
|
zinc finger protein 738
|
|
chr5_-_121975676
Show fit
|
0.56 |
ENSMUST00000197892.3
|
Sh2b3
|
SH2B adaptor protein 3
|
|
chr3_+_90520176
Show fit
|
0.56 |
ENSMUST00000001051.9
|
S100a6
|
S100 calcium binding protein A6 (calcyclin)
|
|
chrX_-_104972938
Show fit
|
0.55 |
ENSMUST00000198448.5
ENSMUST00000199233.5
ENSMUST00000134507.8
ENSMUST00000154866.8
ENSMUST00000128968.8
ENSMUST00000134381.8
ENSMUST00000150914.8
|
Atrx
|
ATRX, chromatin remodeler
|
|
chr8_-_34575946
Show fit
|
0.55 |
ENSMUST00000033913.11
|
Dctn6
|
dynactin 6
|
|
chr9_+_38340751
Show fit
|
0.55 |
ENSMUST00000216502.3
ENSMUST00000216644.2
|
Olfr901
|
olfactory receptor 901
|
|
chr9_+_38725910
Show fit
|
0.55 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922
|
|
chr6_-_48422447
Show fit
|
0.54 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467
|
|
chr3_+_96603694
Show fit
|
0.53 |
ENSMUST00000107076.10
|
Pias3
|
protein inhibitor of activated STAT 3
|
|
chrX_-_163041185
Show fit
|
0.53 |
ENSMUST00000112265.9
|
Bmx
|
BMX non-receptor tyrosine kinase
|
|
chr1_-_131161312
Show fit
|
0.53 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr10_-_86843878
Show fit
|
0.52 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2
|
|
chr14_-_54924382
Show fit
|
0.52 |
ENSMUST00000022793.15
ENSMUST00000111484.9
|
Acin1
|
apoptotic chromatin condensation inducer 1
|
|
chr4_-_148021217
Show fit
|
0.51 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chrX_+_73298388
Show fit
|
0.50 |
ENSMUST00000119197.8
ENSMUST00000088313.5
|
Emd
|
emerin
|
|
chr2_+_34999497
Show fit
|
0.50 |
ENSMUST00000028235.11
ENSMUST00000156933.8
ENSMUST00000028237.15
ENSMUST00000113032.8
|
Cntrl
|
centriolin
|
|
chr17_+_34848507
Show fit
|
0.48 |
ENSMUST00000015620.7
|
Prrt1
|
proline-rich transmembrane protein 1
|
|
chr1_-_169938060
Show fit
|
0.48 |
ENSMUST00000027985.8
ENSMUST00000194690.6
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr5_+_4073343
Show fit
|
0.47 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9
|
|
chr1_+_4878046
Show fit
|
0.47 |
ENSMUST00000027036.11
ENSMUST00000150971.8
ENSMUST00000119612.9
ENSMUST00000137887.8
ENSMUST00000115529.8
|
Lypla1
|
lysophospholipase 1
|
|
chr16_-_3535958
Show fit
|
0.47 |
ENSMUST00000023180.15
|
Mefv
|
Mediterranean fever
|
|
chr10_-_62343516
Show fit
|
0.47 |
ENSMUST00000020271.13
|
Srgn
|
serglycin
|
|
chr7_+_110373447
Show fit
|
0.46 |
ENSMUST00000147587.2
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr18_-_55123153
Show fit
|
0.46 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608
|
|
chr10_-_128246403
Show fit
|
0.46 |
ENSMUST00000164664.8
|
Nabp2
|
nucleic acid binding protein 2
|
|
chr11_+_112673041
Show fit
|
0.46 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9
|
|
chr11_-_78074377
Show fit
|
0.46 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A
|
|
chr17_-_49871291
Show fit
|
0.46 |
ENSMUST00000224595.2
ENSMUST00000057610.8
|
Daam2
|
dishevelled associated activator of morphogenesis 2
|
|
chr10_+_42736345
Show fit
|
0.45 |
ENSMUST00000063063.14
|
Scml4
|
Scm polycomb group protein like 4
|
|
chr9_+_64292957
Show fit
|
0.44 |
ENSMUST00000068967.11
|
Megf11
|
multiple EGF-like-domains 11
|
|
chrX_-_47543029
Show fit
|
0.44 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr9_-_60418286
Show fit
|
0.44 |
ENSMUST00000098660.10
|
Thsd4
|
thrombospondin, type I, domain containing 4
|
|
chr1_+_24717793
Show fit
|
0.42 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1
|
|
chr9_+_64292908
Show fit
|
0.42 |
ENSMUST00000093829.9
ENSMUST00000118485.8
ENSMUST00000164113.8
|
Megf11
|
multiple EGF-like-domains 11
|
|
chr11_-_75918551
Show fit
|
0.42 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B
|
|
chr6_-_88022172
Show fit
|
0.41 |
ENSMUST00000203674.3
ENSMUST00000204126.2
ENSMUST00000113596.8
ENSMUST00000113600.10
|
Rab7
|
RAB7, member RAS oncogene family
|
|
chr16_+_8288627
Show fit
|
0.40 |
ENSMUST00000046470.16
ENSMUST00000150790.2
ENSMUST00000142899.2
|
Mettl22
|
methyltransferase like 22
|
|
chr9_+_124195807
Show fit
|
0.40 |
ENSMUST00000239563.2
|
Ppp2r3d
|
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta
|
|
chr17_+_38082190
Show fit
|
0.39 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122
|
|
chr9_+_65831489
Show fit
|
0.39 |
ENSMUST00000130798.3
|
Csnk1g1
|
casein kinase 1, gamma 1
|
|
chr17_+_3447465
Show fit
|
0.39 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2
|
|
chr9_-_40015750
Show fit
|
0.39 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984
|
|
chr1_+_88139678
Show fit
|
0.38 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chrX_-_63320543
Show fit
|
0.38 |
ENSMUST00000114679.2
ENSMUST00000069926.14
|
Slitrk4
|
SLIT and NTRK-like family, member 4
|