Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
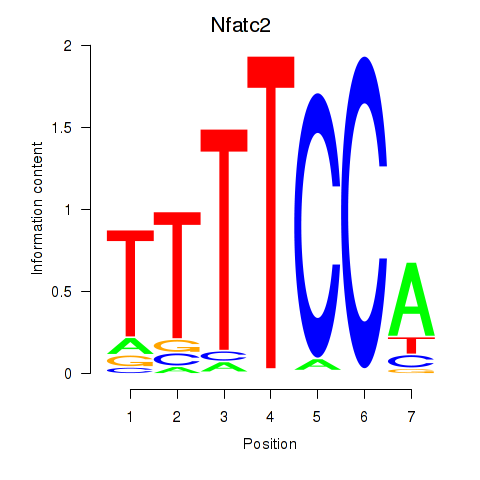
Results for Nfatc2
Z-value: 0.55
Transcription factors associated with Nfatc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc2
|
ENSMUSG00000027544.17 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc2 | mm39_v1_chr2_-_168443540_168443577 | -0.98 | 3.6e-03 | Click! |
Activity profile of Nfatc2 motif
Sorted Z-values of Nfatc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_35587888 | 0.54 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chrX_+_138464065 | 0.36 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr3_-_61273228 | 0.32 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr7_-_126399208 | 0.31 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr14_+_43951187 | 0.30 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr16_-_92196954 | 0.28 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr4_+_102617495 | 0.27 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr1_-_16727133 | 0.26 |
ENSMUST00000185771.7
|
Eloc
|
elongin C |
| chr7_+_100143250 | 0.25 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr7_+_43086432 | 0.25 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr3_+_159545309 | 0.24 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr18_+_60907698 | 0.24 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr7_+_100142977 | 0.24 |
ENSMUST00000129324.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr14_+_74973081 | 0.23 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr6_-_127128007 | 0.23 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr2_-_32602246 | 0.22 |
ENSMUST00000155205.2
ENSMUST00000120105.8 |
Cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr7_+_46496929 | 0.22 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr10_+_53472853 | 0.21 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr12_-_90705212 | 0.21 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr7_+_46496552 | 0.21 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr4_+_102617332 | 0.21 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr17_-_34218301 | 0.20 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr7_+_46496506 | 0.20 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chrX_+_55493325 | 0.20 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr5_+_117501557 | 0.19 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr5_-_121025645 | 0.19 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr18_+_60907668 | 0.19 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr11_-_87878301 | 0.19 |
ENSMUST00000020775.9
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr12_+_75355082 | 0.19 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr3_+_106020545 | 0.18 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr19_-_60770628 | 0.18 |
ENSMUST00000238125.2
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr10_+_7543260 | 0.18 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr19_+_24853039 | 0.18 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr1_+_52026296 | 0.18 |
ENSMUST00000168302.8
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr10_+_4561974 | 0.18 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chrX_-_17437801 | 0.18 |
ENSMUST00000177213.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr7_+_43086554 | 0.18 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr9_+_107445101 | 0.17 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr14_-_31503869 | 0.17 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr4_+_148025316 | 0.17 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr11_+_48977852 | 0.17 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr14_-_65830463 | 0.16 |
ENSMUST00000225355.2
ENSMUST00000022609.7 |
Elp3
|
elongator acetyltransferase complex subunit 3 |
| chr5_+_129864044 | 0.16 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr6_-_16898440 | 0.16 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr18_+_47245204 | 0.16 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr9_+_113641615 | 0.16 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr8_+_3565401 | 0.16 |
ENSMUST00000207146.2
ENSMUST00000208002.2 |
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr1_+_87998487 | 0.16 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr5_+_31070739 | 0.16 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chrX_+_73348832 | 0.16 |
ENSMUST00000153141.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr6_+_88442391 | 0.16 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr5_-_21087023 | 0.15 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr10_-_37014859 | 0.15 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr11_-_48836975 | 0.15 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr4_+_140428777 | 0.15 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr15_-_98851566 | 0.15 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr7_-_99508117 | 0.15 |
ENSMUST00000209032.2
ENSMUST00000036274.8 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr5_+_110478558 | 0.15 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr2_-_151586063 | 0.15 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr7_-_110673269 | 0.15 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr15_-_77037972 | 0.15 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr10_-_119075910 | 0.15 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr15_+_8138805 | 0.15 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr11_+_98938137 | 0.15 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr11_-_75313350 | 0.15 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr10_-_127147609 | 0.15 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr2_-_93292257 | 0.15 |
ENSMUST00000150508.8
|
Cd82
|
CD82 antigen |
| chr1_+_171041539 | 0.15 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr18_+_35347983 | 0.15 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr18_-_43610829 | 0.14 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr12_-_102724931 | 0.14 |
ENSMUST00000174651.2
|
Gm20604
|
predicted gene 20604 |
| chr19_-_6885657 | 0.14 |
ENSMUST00000149261.8
|
Prdx5
|
peroxiredoxin 5 |
| chr6_+_145091196 | 0.14 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr6_+_72575458 | 0.14 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr7_-_99508066 | 0.14 |
ENSMUST00000208477.2
ENSMUST00000208465.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr8_+_3565377 | 0.14 |
ENSMUST00000111070.4
ENSMUST00000004681.14 ENSMUST00000208310.2 |
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr13_-_97897139 | 0.14 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr11_-_80030735 | 0.14 |
ENSMUST00000136996.2
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr11_+_23234644 | 0.14 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr4_+_97665843 | 0.14 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr16_+_32427738 | 0.14 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr13_-_6698751 | 0.14 |
ENSMUST00000021614.14
ENSMUST00000138703.8 |
Pfkp
|
phosphofructokinase, platelet |
| chr7_-_28649094 | 0.13 |
ENSMUST00000148196.3
|
Actn4
|
actinin alpha 4 |
| chr11_+_79883885 | 0.13 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr14_+_8348779 | 0.13 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chrX_+_105230706 | 0.13 |
ENSMUST00000081593.13
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr16_-_37360097 | 0.13 |
ENSMUST00000023525.9
|
Gtf2e1
|
general transcription factor II E, polypeptide 1 (alpha subunit) |
| chr7_-_112946481 | 0.13 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr16_-_30152708 | 0.13 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr17_-_35827676 | 0.13 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr15_+_4055865 | 0.13 |
ENSMUST00000110690.9
|
Oxct1
|
3-oxoacid CoA transferase 1 |
| chr2_+_4022537 | 0.13 |
ENSMUST00000177457.8
|
Frmd4a
|
FERM domain containing 4A |
| chr10_+_80765900 | 0.13 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_-_115426618 | 0.13 |
ENSMUST00000121185.8
ENSMUST00000117589.8 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr14_-_101846459 | 0.13 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr2_+_19376447 | 0.13 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr17_+_28910302 | 0.13 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr6_-_85114725 | 0.13 |
ENSMUST00000174769.2
ENSMUST00000174286.3 ENSMUST00000045986.8 |
Spr
|
sepiapterin reductase |
| chr11_+_48977888 | 0.13 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr12_-_55045887 | 0.13 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chrX_+_93679671 | 0.12 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chrX_-_50106844 | 0.12 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr2_+_21210527 | 0.12 |
ENSMUST00000054591.10
ENSMUST00000102952.8 ENSMUST00000138965.8 ENSMUST00000138914.8 ENSMUST00000102951.2 |
Thnsl1
|
threonine synthase-like 1 (bacterial) |
| chrX_+_152506577 | 0.12 |
ENSMUST00000140575.8
ENSMUST00000208373.2 ENSMUST00000185492.7 ENSMUST00000149514.8 |
Nbdy
|
negative regulator of P-body association |
| chr4_-_154983533 | 0.12 |
ENSMUST00000030935.10
ENSMUST00000132281.2 |
Prxl2b
|
peroxiredoxin like 2B |
| chr6_+_95094721 | 0.12 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr4_-_3835595 | 0.12 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr5_-_137074880 | 0.12 |
ENSMUST00000144303.2
ENSMUST00000111080.8 |
Ap1s1
|
adaptor protein complex AP-1, sigma 1 |
| chr7_+_130179063 | 0.12 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_86148379 | 0.12 |
ENSMUST00000132024.8
ENSMUST00000139285.8 |
Ints2
|
integrator complex subunit 2 |
| chr12_-_79239022 | 0.12 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr11_-_83421333 | 0.12 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr3_-_116474909 | 0.12 |
ENSMUST00000140672.3
|
Gm43191
|
predicted gene 43191 |
| chr3_+_94882142 | 0.12 |
ENSMUST00000167008.8
ENSMUST00000107251.9 |
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr6_-_134864731 | 0.12 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr14_+_71011744 | 0.12 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr9_+_56344700 | 0.12 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr17_+_35113490 | 0.12 |
ENSMUST00000052778.10
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr17_-_28126679 | 0.12 |
ENSMUST00000025057.5
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr5_-_124492734 | 0.12 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr7_-_5128936 | 0.12 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr3_-_107876477 | 0.11 |
ENSMUST00000004136.10
|
Gstm3
|
glutathione S-transferase, mu 3 |
| chr14_-_52248324 | 0.11 |
ENSMUST00000226964.2
|
Zfp219
|
zinc finger protein 219 |
| chr17_+_34423054 | 0.11 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr16_-_35891739 | 0.11 |
ENSMUST00000231351.2
ENSMUST00000004057.9 |
Fam162a
|
family with sequence similarity 162, member A |
| chr4_-_129452180 | 0.11 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr1_+_177272297 | 0.11 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr17_-_37269330 | 0.11 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr15_+_102381929 | 0.11 |
ENSMUST00000229746.2
ENSMUST00000230211.2 ENSMUST00000230539.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr10_-_40178182 | 0.11 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr16_+_32238520 | 0.11 |
ENSMUST00000014220.15
ENSMUST00000080316.8 |
Dynlt2b
|
dynein light chain Tctex-type 2B |
| chr3_-_34135408 | 0.11 |
ENSMUST00000117223.4
ENSMUST00000120805.8 ENSMUST00000011029.12 ENSMUST00000108195.10 |
Dnajc19
|
DnaJ heat shock protein family (Hsp40) member C19 |
| chr11_-_58829738 | 0.11 |
ENSMUST00000094151.6
|
Rnf187
|
ring finger protein 187 |
| chr9_-_108455899 | 0.11 |
ENSMUST00000068700.7
|
Wdr6
|
WD repeat domain 6 |
| chr11_+_93935021 | 0.11 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr9_-_105398346 | 0.11 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr7_-_79882313 | 0.11 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr17_-_29483075 | 0.11 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr15_+_102379503 | 0.11 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr10_+_76304700 | 0.11 |
ENSMUST00000170795.3
|
Mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr12_+_87490666 | 0.11 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr14_+_105496008 | 0.11 |
ENSMUST00000181969.8
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr18_-_36903237 | 0.11 |
ENSMUST00000235494.2
|
Hars
|
histidyl-tRNA synthetase |
| chrX_+_41241049 | 0.11 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr3_-_37180093 | 0.11 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr16_+_93404719 | 0.11 |
ENSMUST00000039659.9
ENSMUST00000231762.2 |
Cbr1
|
carbonyl reductase 1 |
| chr15_+_90108480 | 0.11 |
ENSMUST00000100309.3
ENSMUST00000231200.2 |
Alg10b
|
asparagine-linked glycosylation 10B (alpha-1,2-glucosyltransferase) |
| chr6_-_127127959 | 0.11 |
ENSMUST00000201637.2
|
Ccnd2
|
cyclin D2 |
| chr11_+_93935066 | 0.11 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr3_+_84859453 | 0.11 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr8_+_129450766 | 0.10 |
ENSMUST00000149116.2
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta) |
| chr9_-_64633865 | 0.10 |
ENSMUST00000168366.2
|
Rab11a
|
RAB11A, member RAS oncogene family |
| chr10_-_53952686 | 0.10 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr13_+_83652352 | 0.10 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr14_+_52222283 | 0.10 |
ENSMUST00000093813.12
ENSMUST00000100639.11 ENSMUST00000182909.8 ENSMUST00000182760.8 ENSMUST00000182061.8 ENSMUST00000182193.2 |
Arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr12_+_55883101 | 0.10 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr6_-_127127993 | 0.10 |
ENSMUST00000201066.2
|
Ccnd2
|
cyclin D2 |
| chr17_-_35100980 | 0.10 |
ENSMUST00000152417.8
ENSMUST00000146299.8 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr11_-_93776580 | 0.10 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component |
| chr16_-_45975440 | 0.10 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr15_-_89007290 | 0.10 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr17_+_45817750 | 0.10 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr8_+_22901377 | 0.10 |
ENSMUST00000033934.5
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr17_+_6307123 | 0.10 |
ENSMUST00000232383.2
|
Tmem181a
|
transmembrane protein 181A |
| chr1_+_177272215 | 0.10 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chrX_-_133501874 | 0.10 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr5_+_108213608 | 0.10 |
ENSMUST00000081567.11
ENSMUST00000170319.8 ENSMUST00000112626.8 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr3_-_19319155 | 0.10 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr12_-_65010124 | 0.10 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr5_+_86219593 | 0.10 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr7_-_132178101 | 0.10 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr1_-_156501860 | 0.10 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr2_-_92876398 | 0.10 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chr17_-_37269153 | 0.10 |
ENSMUST00000172823.2
|
Polr1h
|
RNA polymerase I subunit H |
| chr3_+_88523730 | 0.10 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr17_-_37269425 | 0.10 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr16_+_4501934 | 0.10 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr11_-_69906171 | 0.09 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr11_-_17161504 | 0.09 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chrX_-_133501677 | 0.09 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr4_-_129452148 | 0.09 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr10_-_93146825 | 0.09 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr11_+_29642937 | 0.09 |
ENSMUST00000102843.10
ENSMUST00000102842.10 ENSMUST00000078830.11 ENSMUST00000170731.8 |
Rtn4
|
reticulon 4 |
| chr12_+_102724223 | 0.09 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr14_+_54924439 | 0.09 |
ENSMUST00000227269.2
|
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr1_-_37575313 | 0.09 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr17_-_36149100 | 0.09 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr14_+_105496147 | 0.09 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr3_+_109481223 | 0.09 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr5_-_124616393 | 0.09 |
ENSMUST00000031347.8
|
Rilpl2
|
Rab interacting lysosomal protein-like 2 |
| chr7_-_79882228 | 0.09 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr2_+_78699360 | 0.09 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_-_115236354 | 0.09 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr5_+_44070486 | 0.09 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr1_-_16727242 | 0.09 |
ENSMUST00000186948.7
ENSMUST00000187910.7 ENSMUST00000115352.10 |
Eloc
|
elongin C |
| chr1_-_16727067 | 0.09 |
ENSMUST00000188641.7
|
Eloc
|
elongin C |
| chrX_+_133501928 | 0.09 |
ENSMUST00000074950.11
ENSMUST00000113203.2 ENSMUST00000113202.8 ENSMUST00000050331.13 ENSMUST00000059297.6 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr1_-_172125555 | 0.09 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr15_+_102381705 | 0.09 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr14_+_79753055 | 0.09 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.3 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.6 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.2 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.0 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.0 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.0 | 0.1 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0097017 | renal protein absorption(GO:0097017) positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0048003 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.1 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0061087 | regulation of histone H3-K27 methylation(GO:0061085) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.2 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.2 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0098837 | kinetochore microtubule(GO:0005828) postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.2 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0036461 | AP-3 adaptor complex binding(GO:0035651) BLOC-2 complex binding(GO:0036461) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |