Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
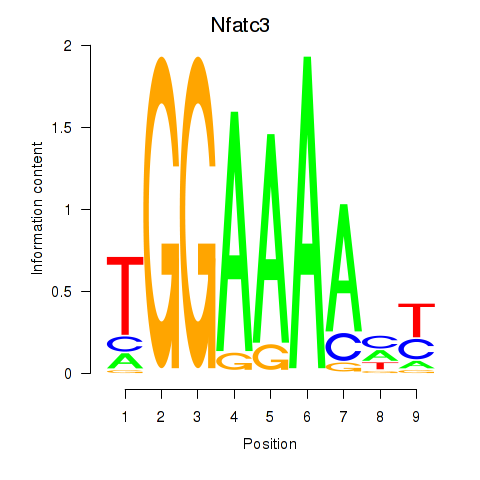
Results for Nfatc3
Z-value: 2.25
Transcription factors associated with Nfatc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc3
|
ENSMUSG00000031902.11 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc3 | mm39_v1_chr8_+_106786190_106786265 | 1.00 | 3.3e-04 | Click! |
Activity profile of Nfatc3 motif
Sorted Z-values of Nfatc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_142326363 | 1.62 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr10_+_4561974 | 1.48 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr11_+_48977852 | 1.45 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr7_+_46496929 | 1.42 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr7_+_46496552 | 1.35 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr7_+_46496506 | 1.31 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr7_+_100143250 | 1.24 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr5_-_88675700 | 1.20 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr9_-_105398346 | 1.18 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr3_-_61273228 | 1.17 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr2_-_32602246 | 1.16 |
ENSMUST00000155205.2
ENSMUST00000120105.8 |
Cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr7_+_100142977 | 1.14 |
ENSMUST00000129324.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_+_98938137 | 1.13 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr11_+_48977888 | 1.02 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chrX_+_138464065 | 1.02 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr10_+_7543260 | 0.92 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr9_+_88209250 | 0.86 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr8_+_85696695 | 0.80 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr14_+_43951187 | 0.79 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr8_+_85696453 | 0.78 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr5_+_117501557 | 0.78 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr15_+_4055865 | 0.77 |
ENSMUST00000110690.9
|
Oxct1
|
3-oxoacid CoA transferase 1 |
| chr15_-_98851566 | 0.70 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr8_+_85696396 | 0.70 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_3565401 | 0.69 |
ENSMUST00000207146.2
ENSMUST00000208002.2 |
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr6_+_30401864 | 0.69 |
ENSMUST00000068240.13
ENSMUST00000068259.10 ENSMUST00000132581.8 |
Klhdc10
|
kelch domain containing 10 |
| chr1_-_93373145 | 0.66 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr14_+_8348779 | 0.66 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr7_-_28649094 | 0.64 |
ENSMUST00000148196.3
|
Actn4
|
actinin alpha 4 |
| chr15_+_8138805 | 0.63 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr11_-_3864664 | 0.63 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr5_-_148329615 | 0.63 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr14_+_79753055 | 0.62 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr14_+_55797443 | 0.61 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_-_80030735 | 0.61 |
ENSMUST00000136996.2
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr12_+_52550775 | 0.61 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr8_+_3565377 | 0.60 |
ENSMUST00000111070.4
ENSMUST00000004681.14 ENSMUST00000208310.2 |
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr19_-_42741148 | 0.60 |
ENSMUST00000236659.2
ENSMUST00000076505.4 |
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr9_-_99450948 | 0.60 |
ENSMUST00000035043.12
|
Armc8
|
armadillo repeat containing 8 |
| chr17_+_73144531 | 0.58 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr5_+_110478558 | 0.57 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr17_-_35827676 | 0.56 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr13_+_49735938 | 0.56 |
ENSMUST00000221170.2
|
Omd
|
osteomodulin |
| chr10_+_80905869 | 0.56 |
ENSMUST00000005057.7
|
Thop1
|
thimet oligopeptidase 1 |
| chrX_-_153999440 | 0.56 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr2_-_25498459 | 0.56 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr3_+_89987749 | 0.56 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr3_-_37180093 | 0.56 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr3_+_159545309 | 0.56 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr5_-_21087023 | 0.55 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr13_-_6698751 | 0.55 |
ENSMUST00000021614.14
ENSMUST00000138703.8 |
Pfkp
|
phosphofructokinase, platelet |
| chr12_-_55045887 | 0.54 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr3_-_94566107 | 0.53 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr7_-_45480200 | 0.53 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr8_-_85696369 | 0.53 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr5_+_44070486 | 0.53 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr19_+_24853039 | 0.52 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr6_+_18848570 | 0.51 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr17_-_45903410 | 0.51 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr2_+_21210527 | 0.49 |
ENSMUST00000054591.10
ENSMUST00000102952.8 ENSMUST00000138965.8 ENSMUST00000138914.8 ENSMUST00000102951.2 |
Thnsl1
|
threonine synthase-like 1 (bacterial) |
| chr1_+_152683568 | 0.49 |
ENSMUST00000190323.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chrX_-_8059597 | 0.49 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr1_+_87998487 | 0.49 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr6_+_18848600 | 0.49 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr12_+_31704853 | 0.48 |
ENSMUST00000036862.5
|
Cog5
|
component of oligomeric golgi complex 5 |
| chr14_-_75991903 | 0.48 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr3_+_84859453 | 0.48 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr7_+_130179063 | 0.47 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr9_+_65370077 | 0.47 |
ENSMUST00000215170.2
|
Spg21
|
SPG21, maspardin |
| chr12_-_31704766 | 0.47 |
ENSMUST00000020977.4
|
Dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr10_+_61516078 | 0.47 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr15_+_41694317 | 0.47 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr15_+_102381705 | 0.47 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr11_+_95557783 | 0.47 |
ENSMUST00000125172.8
ENSMUST00000036374.6 |
Phb
|
prohibitin |
| chr10_-_57408512 | 0.46 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr10_-_53952686 | 0.46 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr9_+_54858388 | 0.46 |
ENSMUST00000171900.2
|
Psma4
|
proteasome subunit alpha 4 |
| chr8_-_46605196 | 0.45 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr1_-_106641940 | 0.45 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr7_+_27186335 | 0.45 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr7_-_126873219 | 0.45 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr1_-_16689660 | 0.45 |
ENSMUST00000117146.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr15_-_98432111 | 0.45 |
ENSMUST00000116400.4
ENSMUST00000023727.17 ENSMUST00000230542.2 |
Kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr2_+_61542038 | 0.44 |
ENSMUST00000028278.14
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr10_+_53472853 | 0.44 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr9_+_56344700 | 0.44 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr12_-_85335193 | 0.43 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr3_+_88523730 | 0.43 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr16_-_19801781 | 0.42 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr4_-_129452180 | 0.42 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr17_-_37269330 | 0.42 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr11_+_109376432 | 0.42 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr7_-_126073994 | 0.41 |
ENSMUST00000205733.2
ENSMUST00000205889.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chrX_-_153999333 | 0.41 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr7_-_126074222 | 0.41 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr7_-_103928939 | 0.40 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr13_+_93441307 | 0.40 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr7_-_5128936 | 0.40 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr16_-_91723870 | 0.40 |
ENSMUST00000159295.8
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr15_-_66703471 | 0.40 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr4_-_129452148 | 0.40 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr19_+_56414114 | 0.39 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr18_+_69633741 | 0.39 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr3_+_87432879 | 0.39 |
ENSMUST00000170036.8
ENSMUST00000117293.2 |
Etv3
|
ets variant 3 |
| chr6_+_127423779 | 0.39 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr14_+_19801333 | 0.39 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr5_+_33815466 | 0.39 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr16_+_35892437 | 0.38 |
ENSMUST00000163352.9
ENSMUST00000231468.2 |
Ccdc58
|
coiled-coil domain containing 58 |
| chr2_-_125701059 | 0.38 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr13_+_69950509 | 0.37 |
ENSMUST00000223376.2
ENSMUST00000222387.2 |
Med10
|
mediator complex subunit 10 |
| chr9_+_96140781 | 0.37 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr1_-_180021039 | 0.37 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr2_-_181223787 | 0.36 |
ENSMUST00000057816.15
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr8_+_106412905 | 0.36 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr6_-_72357398 | 0.36 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr12_+_111409087 | 0.36 |
ENSMUST00000109792.8
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr17_-_37269425 | 0.36 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr2_+_125701054 | 0.36 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr12_+_4684620 | 0.35 |
ENSMUST00000217672.2
|
Itsn2
|
intersectin 2 |
| chr2_+_35172392 | 0.35 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
| chr1_-_181039509 | 0.34 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr7_-_126074256 | 0.34 |
ENSMUST00000205440.2
ENSMUST00000032978.8 ENSMUST00000205340.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr15_-_83316995 | 0.34 |
ENSMUST00000165095.9
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr8_-_94739469 | 0.34 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr4_+_86493905 | 0.34 |
ENSMUST00000091064.8
|
Rraga
|
Ras-related GTP binding A |
| chr12_+_84332006 | 0.34 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr6_-_71609881 | 0.34 |
ENSMUST00000065509.11
ENSMUST00000207023.2 |
Kdm3a
|
lysine (K)-specific demethylase 3A |
| chr11_+_54517164 | 0.34 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr5_-_88823049 | 0.34 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chrX_+_152506577 | 0.34 |
ENSMUST00000140575.8
ENSMUST00000208373.2 ENSMUST00000185492.7 ENSMUST00000149514.8 |
Nbdy
|
negative regulator of P-body association |
| chr14_-_47059546 | 0.33 |
ENSMUST00000226937.2
|
Gmfb
|
glia maturation factor, beta |
| chr14_+_55797468 | 0.33 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr13_+_93441447 | 0.33 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr1_+_171041539 | 0.33 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chrX_-_47602395 | 0.33 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr18_+_67338437 | 0.33 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr2_-_125700905 | 0.33 |
ENSMUST00000110462.8
|
Cops2
|
COP9 signalosome subunit 2 |
| chr15_-_10470575 | 0.33 |
ENSMUST00000136591.8
|
Dnajc21
|
DnaJ heat shock protein family (Hsp40) member C21 |
| chr15_-_83317020 | 0.32 |
ENSMUST00000231184.2
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_-_123126550 | 0.32 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_-_23821523 | 0.32 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr4_+_155896946 | 0.32 |
ENSMUST00000030944.11
|
Ccnl2
|
cyclin L2 |
| chr12_+_10440755 | 0.32 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr6_+_54249817 | 0.31 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr8_-_84831391 | 0.31 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr14_-_55897854 | 0.31 |
ENSMUST00000002400.7
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr3_+_85946145 | 0.31 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chrX_-_133501677 | 0.31 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr15_-_100322934 | 0.30 |
ENSMUST00000123461.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr11_-_121095462 | 0.30 |
ENSMUST00000026169.7
|
Ogfod3
|
2-oxoglutarate and iron-dependent oxygenase domain containing 3 |
| chr6_+_29853745 | 0.30 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr1_+_55276336 | 0.30 |
ENSMUST00000061334.10
|
Mars2
|
methionine-tRNA synthetase 2 (mitochondrial) |
| chr13_-_114524586 | 0.30 |
ENSMUST00000232101.2
ENSMUST00000225978.3 |
Ndufs4
|
NADH:ubiquinone oxidoreductase core subunit S4 |
| chr17_+_36152559 | 0.30 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr3_-_51468236 | 0.29 |
ENSMUST00000037141.9
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr6_-_51443602 | 0.29 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr4_-_63779562 | 0.29 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr6_-_129449739 | 0.29 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr5_-_105198913 | 0.29 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr17_-_45903494 | 0.29 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr13_+_25127127 | 0.28 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr4_+_43406435 | 0.28 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr7_-_99994257 | 0.28 |
ENSMUST00000207634.2
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr13_+_16189041 | 0.28 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chrX_-_133501874 | 0.28 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr18_+_24087725 | 0.27 |
ENSMUST00000225682.2
ENSMUST00000060762.6 |
Zfp397
|
zinc finger protein 397 |
| chr2_+_23046381 | 0.27 |
ENSMUST00000028117.4
|
Yme1l1
|
YME1-like 1 (S. cerevisiae) |
| chr7_-_130148984 | 0.27 |
ENSMUST00000160289.9
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr17_+_28988354 | 0.26 |
ENSMUST00000233109.2
ENSMUST00000004986.14 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr11_+_48977495 | 0.26 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr17_+_6320731 | 0.26 |
ENSMUST00000088940.6
|
Tmem181a
|
transmembrane protein 181A |
| chr5_+_3393893 | 0.26 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr6_+_3498382 | 0.26 |
ENSMUST00000001412.17
ENSMUST00000170873.10 ENSMUST00000164052.5 |
Vps50
|
VPS50 EARP/GARPII complex subunit |
| chr14_-_72840373 | 0.26 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr7_-_126483851 | 0.26 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr9_-_37166699 | 0.26 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr5_-_130284366 | 0.26 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr10_-_44334711 | 0.26 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr11_-_83421333 | 0.25 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr15_-_91075933 | 0.25 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr14_-_52248324 | 0.24 |
ENSMUST00000226964.2
|
Zfp219
|
zinc finger protein 219 |
| chr18_+_37858753 | 0.24 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr6_+_134807097 | 0.24 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr6_+_71350411 | 0.24 |
ENSMUST00000066747.14
|
Cd8a
|
CD8 antigen, alpha chain |
| chr6_+_48818567 | 0.23 |
ENSMUST00000203639.3
|
Tmem176a
|
transmembrane protein 176A |
| chr19_-_6947112 | 0.23 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chrX_-_101687813 | 0.23 |
ENSMUST00000052012.14
ENSMUST00000043596.12 ENSMUST00000119229.8 ENSMUST00000122022.8 ENSMUST00000120270.8 ENSMUST00000113611.3 |
Phka1
|
phosphorylase kinase alpha 1 |
| chr11_-_107238956 | 0.23 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_+_56905698 | 0.23 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr17_+_29898221 | 0.22 |
ENSMUST00000129864.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr16_+_43960183 | 0.22 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr17_-_52133594 | 0.22 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr1_-_16689527 | 0.22 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr3_-_53370621 | 0.22 |
ENSMUST00000056749.14
|
Nhlrc3
|
NHL repeat containing 3 |
| chr8_-_46577183 | 0.22 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr9_+_118307412 | 0.22 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr17_+_26780486 | 0.21 |
ENSMUST00000001619.16
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr12_-_101784727 | 0.21 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr10_-_118705029 | 0.21 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr16_-_43959559 | 0.20 |
ENSMUST00000063661.13
ENSMUST00000114666.9 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 | 1.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 1.2 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 0.9 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.2 | 1.0 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 2.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 | 0.6 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.2 | 0.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 0.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 0.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.4 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 2.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.1 | 0.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.5 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 0.3 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.5 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.5 | GO:0032847 | cochlear nucleus development(GO:0021747) regulation of cellular pH reduction(GO:0032847) |
| 0.1 | 0.5 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.4 | GO:1903921 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.3 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.1 | 0.6 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 4.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.4 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.2 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 1.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.8 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.8 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.0 | 0.2 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.3 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.4 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.5 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.1 | GO:0061295 | regulation of pronephros size(GO:0035565) hepatoblast differentiation(GO:0061017) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.7 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 1.1 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.0 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 4.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.3 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.1 | 1.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 1.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 2.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 1.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 2.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 4.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 0.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 0.6 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.2 | 0.8 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 0.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 2.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.3 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.7 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.8 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.3 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0031812 | beta-1 adrenergic receptor binding(GO:0031697) G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 6.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 5.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |