Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
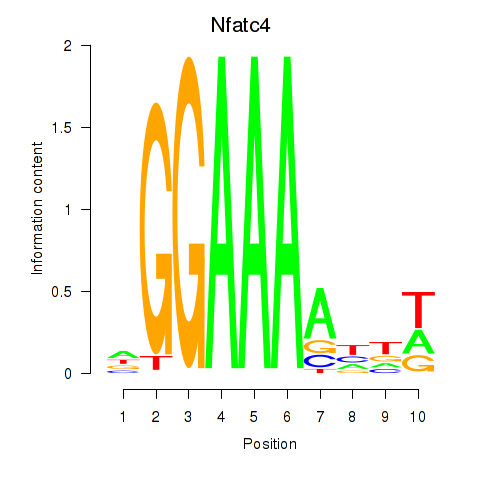
Results for Nfatc4
Z-value: 0.71
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSMUSG00000023411.13 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc4 | mm39_v1_chr14_+_56062422_56062494 | 0.55 | 3.4e-01 | Click! |
Activity profile of Nfatc4 motif
Sorted Z-values of Nfatc4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_129834788 | 0.46 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr2_-_73284262 | 0.45 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_34044940 | 0.39 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr4_+_132903646 | 0.36 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr19_+_8828132 | 0.34 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr15_+_3300249 | 0.32 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr12_+_76580386 | 0.32 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr19_+_26600820 | 0.31 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_150341911 | 0.31 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr19_-_24454720 | 0.29 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr9_+_77543776 | 0.26 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr10_-_12689345 | 0.22 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr3_-_96501443 | 0.22 |
ENSMUST00000145001.2
ENSMUST00000091924.10 |
Polr3gl
|
polymerase (RNA) III (DNA directed) polypeptide G like |
| chrX_-_7537580 | 0.21 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr15_-_79816785 | 0.18 |
ENSMUST00000089293.11
ENSMUST00000109616.9 |
Cbx7
|
chromobox 7 |
| chr8_+_129085719 | 0.16 |
ENSMUST00000026917.10
|
Nrp1
|
neuropilin 1 |
| chr14_+_65187485 | 0.15 |
ENSMUST00000043914.8
ENSMUST00000239450.2 |
Ints9
|
integrator complex subunit 9 |
| chr5_+_53747796 | 0.15 |
ENSMUST00000113865.5
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_174171799 | 0.14 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr14_-_79718890 | 0.12 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr9_-_64858878 | 0.12 |
ENSMUST00000037798.8
|
Slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr15_-_79816717 | 0.12 |
ENSMUST00000177044.2
ENSMUST00000109615.8 |
Cbx7
|
chromobox 7 |
| chr19_-_8816530 | 0.11 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr14_-_65187287 | 0.10 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr1_-_156766381 | 0.10 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_+_51959534 | 0.10 |
ENSMUST00000061352.11
|
Rdx
|
radixin |
| chr10_+_23727325 | 0.10 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr2_+_174171860 | 0.10 |
ENSMUST00000109087.8
ENSMUST00000109084.8 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr3_+_94744844 | 0.09 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr3_-_20329823 | 0.09 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr6_+_134958681 | 0.07 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr1_-_169938060 | 0.07 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr19_+_8816663 | 0.07 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr5_+_53747556 | 0.07 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr17_-_49871291 | 0.07 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_+_91367764 | 0.06 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr4_-_148244299 | 0.06 |
ENSMUST00000151127.8
ENSMUST00000105705.9 |
Fbxo44
|
F-box protein 44 |
| chr4_-_3938352 | 0.06 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr3_-_86455575 | 0.06 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr4_+_48049080 | 0.05 |
ENSMUST00000153369.2
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr19_-_4993060 | 0.05 |
ENSMUST00000133504.2
ENSMUST00000133254.2 ENSMUST00000120475.8 ENSMUST00000025834.15 |
Peli3
|
pellino 3 |
| chr16_+_43184507 | 0.05 |
ENSMUST00000148775.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_156681991 | 0.05 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr13_+_83652150 | 0.04 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr14_+_120513106 | 0.04 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr14_+_120513076 | 0.03 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr2_-_165726610 | 0.03 |
ENSMUST00000177633.8
ENSMUST00000018050.14 ENSMUST00000088113.11 |
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr12_-_70033732 | 0.03 |
ENSMUST00000021467.8
|
Sav1
|
salvador family WW domain containing 1 |
| chr12_-_84497718 | 0.03 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr2_+_156681927 | 0.03 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr6_-_128599833 | 0.02 |
ENSMUST00000171306.5
ENSMUST00000204819.3 ENSMUST00000032512.15 |
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr12_+_84498196 | 0.02 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr18_+_73996743 | 0.02 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr19_-_13080648 | 0.02 |
ENSMUST00000076729.4
|
Olfr1458
|
olfactory receptor 1458 |
| chr16_+_38182569 | 0.02 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr14_+_53980561 | 0.02 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr2_+_55325931 | 0.02 |
ENSMUST00000067101.10
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr14_+_70793340 | 0.02 |
ENSMUST00000163060.2
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr11_-_35871300 | 0.01 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr12_-_75463555 | 0.01 |
ENSMUST00000051079.4
|
Gphb5
|
glycoprotein hormone beta 5 |
| chr9_+_107869662 | 0.01 |
ENSMUST00000177173.8
|
Cdhr4
|
cadherin-related family member 4 |
| chr11_+_75358866 | 0.01 |
ENSMUST00000043598.14
ENSMUST00000108435.2 |
Tlcd2
|
TLC domain containing 2 |
| chr7_+_80764564 | 0.01 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr2_+_36827133 | 0.01 |
ENSMUST00000095021.2
|
Olfr356
|
olfactory receptor 356 |
| chrX_+_3076875 | 0.01 |
ENSMUST00000189190.2
|
Btbd35f23
|
BTB domain containing 35, family member 23 |
| chr17_+_48037758 | 0.01 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr13_+_83652280 | 0.00 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_-_47270201 | 0.00 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr7_+_80764547 | 0.00 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr2_+_84867783 | 0.00 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chrX_+_56144946 | 0.00 |
ENSMUST00000141936.2
|
Vgll1
|
vestigial like family member 1 |
| chrX_-_93408176 | 0.00 |
ENSMUST00000239046.2
|
CXorf58
|
family with sequence similarity 90, member A1B |
| chr8_-_39128662 | 0.00 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr14_+_53284744 | 0.00 |
ENSMUST00000103606.2
|
Trav16d-dv11
|
T cell receptor alpha variable 16D-DV11 |
| chr2_-_73490719 | 0.00 |
ENSMUST00000154258.8
|
Chn1
|
chimerin 1 |
| chr2_+_3115250 | 0.00 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0038190 | cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.2 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:1900625 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |